3gfp
From Proteopedia
m (Protected "3gfp" [edit=sysop:move=sysop]) |
|||
| Line 1: | Line 1: | ||
[[Image:3gfp.png|left|200px]] | [[Image:3gfp.png|left|200px]] | ||
| - | <!-- | ||
| - | The line below this paragraph, containing "STRUCTURE_3gfp", creates the "Structure Box" on the page. | ||
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet) | ||
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | ||
| - | or leave the SCENE parameter empty for the default display. | ||
| - | --> | ||
{{STRUCTURE_3gfp| PDB=3gfp | SCENE= }} | {{STRUCTURE_3gfp| PDB=3gfp | SCENE= }} | ||
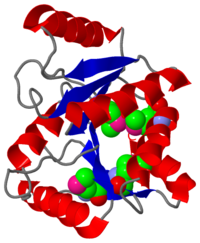
===Structure of the C-terminal domain of the DEAD-box protein Dbp5=== | ===Structure of the C-terminal domain of the DEAD-box protein Dbp5=== | ||
| - | |||
| - | <!-- | ||
| - | The line below this paragraph, {{ABSTRACT_PUBMED_19805289}}, adds the Publication Abstract to the page | ||
| - | (as it appears on PubMed at http://www.pubmed.gov), where 19805289 is the PubMed ID number. | ||
| - | --> | ||
{{ABSTRACT_PUBMED_19805289}} | {{ABSTRACT_PUBMED_19805289}} | ||
| Line 22: | Line 11: | ||
==See Also== | ==See Also== | ||
| + | *[[C-terminal domain of the DEAD-box protein Dbp5|C-terminal domain of the DEAD-box protein Dbp5]] | ||
*[[Helicase|Helicase]] | *[[Helicase|Helicase]] | ||
| - | *[[User:Wayne Decatur|User:Wayne Decatur]] | ||
==Reference== | ==Reference== | ||
Revision as of 06:45, 27 July 2012
Contents |
Structure of the C-terminal domain of the DEAD-box protein Dbp5
The DExD/H-box RNA-dependent ATPase Dbp5 plays an essential role in the nuclear export of mRNA. Dbp5 localizes to the nuclear pore complex, where its ATPase activity is stimulated by Gle1 and its coactivator inositol hexakisphosphate. Here, we present the crystal structure of the C-terminal domain of Dbp5, refined to 1.8 A. The structure reveals a RecA-like fold that contains two defining characteristics not present in other structurally characterized DExD/H-box proteins: a C-terminal alpha-helix and a loop connecting beta5 and alpha4, both of which are composed of conserved and unique elements in the Dbp5 primary sequence. Using structure-guided mutagenesis, we have identified several charged surface residues that, when mutated, weaken the binding of Gle1 and inhibit the ability of Gle1 to stimulate Dbp5's ATPase activity. In vivo analysis of the same mutations reveals that those mutants displaying the weakest ATPase stimulation in vitro are also unable to support yeast growth. Analysis of the correlation between the in vitro and in vivo data indicates that a threshold level of Dbp5 ATPase activity is required for cellular mRNA export that is not met by the unstimulated enzyme, suggesting a possible mechanism by which Dbp5's activity can be modulated to regulate mRNA export.
Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1., Dossani ZY, Weirich CS, Erzberger JP, Berger JM, Weis K, Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16251-6. Epub 2009 Sep 2. PMID:19805289
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
3gfp is a 1 chain structure of Helicase with sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
See Also
Reference
- Dossani ZY, Weirich CS, Erzberger JP, Berger JM, Weis K. Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16251-6. Epub 2009 Sep 2. PMID:19805289
Categories: Saccharomyces cerevisiae | Berger, J M. | Dossani, Z Y. | Erzberger, J P. | Weirich, C S. | Weis, K. | Atp-binding | Atpase | Helicase | Hydrolase | Membrane | Mrna export | Mrna transport | Nuclear pore complex | Nucleotide-binding | Nucleus | Phosphoprotein | Protein transport | Reca-fold | Rna-binding | Translocation | Transport