We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 645
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
== HIV-1 Protease == | == HIV-1 Protease == | ||
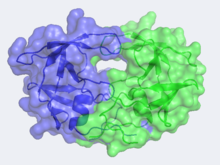
<StructureSection load='3hvp' size='400' side='right' caption='Structure of HIV-1 Protease (PDB entry [[2nmz]])' scene='HIV-1_protease/2nmz/5'> [[Image:CannergreyHIV2.png|220px|left]][[Human Immunodeficiency Virus-1 Protease]] | <StructureSection load='3hvp' size='400' side='right' caption='Structure of HIV-1 Protease (PDB entry [[2nmz]])' scene='HIV-1_protease/2nmz/5'> [[Image:CannergreyHIV2.png|220px|left]][[Human Immunodeficiency Virus-1 Protease]] | ||
| - | + | ||
=='''Introduction'''== | =='''Introduction'''== | ||
| - | HIV -1 protease (HIV PR ) is a retroviral aspartyl protease that is derived from HIV-1, a lentivirus that is best characterized for its ability to lower host immunity by infecting CD4+ T lymphocytes, macrophages, and dendritic cells. Aspartyl proteases are protease enzymes that utilize aspartate residue(s) for the catalysis of peptide substrates. Eukaryotic forms of these proteases include the <scene name='Sandbox_645/Pepsin/3'>pepsins</scene>, cathepsins and renins. While they have a two-domain structure, the retroviral aspartyl proteases are much smaller and homologous to a single domain of the eukaryotic aspartic proteases. | + | HIV -1 protease (HIV PR ) is a retroviral aspartyl protease that is derived from HIV-1, a lentivirus that is best characterized for its ability to lower host immunity by infecting CD4+ T lymphocytes, macrophages, and dendritic cells. Aspartyl proteases are protease enzymes that utilize aspartate residue(s) for the catalysis of peptide substrates. Eukaryotic forms of these proteases include the <scene name='Sandbox_645/Pepsin/3'>pepsins</scene>, cathepsins and renins. While they have a two-domain structure, the retroviral aspartyl proteases are much smaller and homologous to a single domain of the eukaryotic aspartic proteases. |
| + | |||
| + | HIV-1 protease is essential for the life cycle of HIV. The protease takes newly synthesized polyproteins and cleaves them by means of a hydrolysis reaction into the smaller mature protein components of the HIV virion. These proteins are used to form the virion capsid that encases the viral genome. Effectively inhibiting HIV-1 protease will result in the inability for HIV to propagate as it will no longer have the ability to form complete virion units to infect new cells. | ||
| + | |||
| + | |||
| + | The HIV PR, together with single stranded RNA (ssRNA), reverse transcriptase, integrase, and other viral factors, is found inside the HIV-1 virion. As an important viral protein, it plays a crucial role in successful viral propagation. | ||
| Line 9: | Line 14: | ||
Structure of HIV-1 Protease | Structure of HIV-1 Protease | ||
| + | Unlike most members of the aspartyl protease class, which generally exist as two domain monomers, HIV protease is a dimmer with two identical <scene name='Sandbox_645/Monomer/2'>subunits</scene> that are comprised of 99 amino acids. | ||
| + | |||
The X-ray structure of HIV-1 protease reveals that it is composed of <scene name='User:David_Canner/Sandbox_HIV/Identical_subunits/1'>two symmetrically related subunits</scene>, each consisting of 99 amino acid residues. The subunits come together in such as way as to <scene name='User:David_Canner/Sandbox_HIV/Tunnel/1'>form a tunnel where they meet</scene>. This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of <scene name='User:David_Canner/Sandbox_HIV/Catalytic_triad/3'> two Asp-Thr-Gly conserved sequences</scene>, making it a member of the aspartyl protease family. The two Asp's are <scene name='User:David_Canner/Sandbox_HIV/Catalytic_asp/1'>essential catalytic residues</scene> that activate a water molecule to hydrolytically cleave the polyprotein that binds in the tunnel.<ref>PMID:1799632</ref> You may be wondering how a polyprotein makes its way into the active-site tunnel, as the<scene name='User:David_Canner/Sandbox_HIV/Narrow_tunnel/1'> tunnel appears to be too narrow </scene> to admit it. The key is the two flexible flaps on the top of the tunnel that <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph/3'>move to allow proteins </scene>to enter the tunnel. The flaps <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph_flaps/2'>undergo a dramatic movement</scene>, shifting from an open to a closed conformation to bind the target in an appropriate conformation for cleavage. | The X-ray structure of HIV-1 protease reveals that it is composed of <scene name='User:David_Canner/Sandbox_HIV/Identical_subunits/1'>two symmetrically related subunits</scene>, each consisting of 99 amino acid residues. The subunits come together in such as way as to <scene name='User:David_Canner/Sandbox_HIV/Tunnel/1'>form a tunnel where they meet</scene>. This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of <scene name='User:David_Canner/Sandbox_HIV/Catalytic_triad/3'> two Asp-Thr-Gly conserved sequences</scene>, making it a member of the aspartyl protease family. The two Asp's are <scene name='User:David_Canner/Sandbox_HIV/Catalytic_asp/1'>essential catalytic residues</scene> that activate a water molecule to hydrolytically cleave the polyprotein that binds in the tunnel.<ref>PMID:1799632</ref> You may be wondering how a polyprotein makes its way into the active-site tunnel, as the<scene name='User:David_Canner/Sandbox_HIV/Narrow_tunnel/1'> tunnel appears to be too narrow </scene> to admit it. The key is the two flexible flaps on the top of the tunnel that <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph/3'>move to allow proteins </scene>to enter the tunnel. The flaps <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph_flaps/2'>undergo a dramatic movement</scene>, shifting from an open to a closed conformation to bind the target in an appropriate conformation for cleavage. | ||
Revision as of 05:34, 27 November 2012
HIV-1 Protease
| |||||||||||