Sandbox Reserved 592
From Proteopedia
| Line 47: | Line 47: | ||
| | | | ||
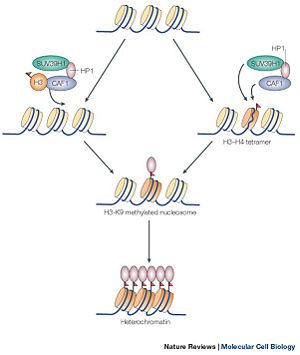
| - | [[Image:picture 3.jpg | 300 px | thumb |Figure | + | [[Image:picture 3.jpg | 300 px | thumb |Figure 3 showing the methylation of the histone protein and the subsequent formation of heterochromatin]] |
---- | ---- | ||
Revision as of 07:05, 23 April 2013
==Your Heading Here (maybe something like 'Structure'-- PLEASE DO NOT DELETE THIS TEMPLATE -->
| This Sandbox is Reserved from Feb 1, 2013, through May 10, 2013 for use in the course "Biochemistry" taught by Irma Santoro at the Reinhardt University. This reservation includes Sandbox Reserved 591 through Sandbox Reserved 599. |
To get started:
More help: Help:Editing |
Contents |
Background
SUV39h1 is part of a class of methytransferase that deals with the methylation of histone proteins in nucleosomes. The methylation of histone proteins is an essential part of an area of genetics which deals with the modification of histone proteins called epigenomics. Epigenomics is the study of modifications of nucleosomes, which either inhibit or express gene transcription without changing the underlining DNA sequence. These changes are allowed because the amino acid tails which extend out and away from the histone proteins, exposing itself to methylation. Methylation of histone protein is one of many ways that regulates gene expression. The regulation of gene expression is dependent on the state of the gene. A gene cannot be transcribed when the promoter of the gene is wrapped around the nucleosome, forming a heterochromatin state; thus, the gene is inhibited from being transcribed. coversly Methylation of the histone protein causes the winding around the nucleosomes, transforming a euchromatin state into a heterochromatin state. Of course, SUV39H1 alone does not methtylate the histone proteins. SUV39H1 interacts with other proteins in order to efficiently methylate a histone protein (shown in Figure 1).