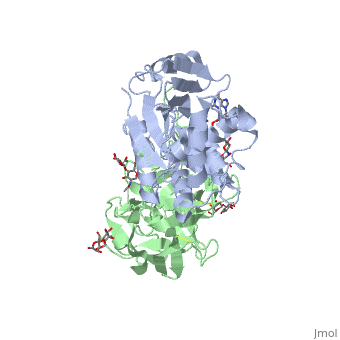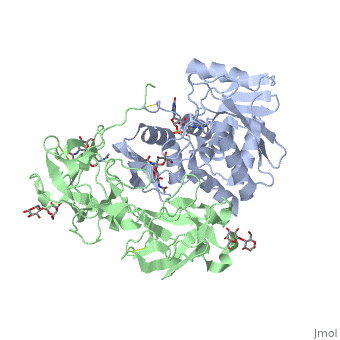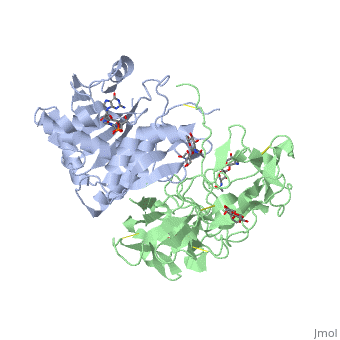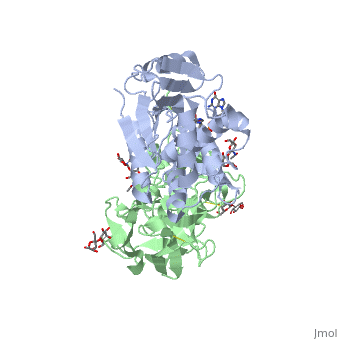
Sandbox BCMB402 Ricin
From Proteopedia
m |
|||
| Line 12: | Line 12: | ||
| - | The B chain is a lectin<ref name="lord" /> that binds to galactose-containing surface receptors | + | The B chain is a lectin<ref name="lord" /> that binds to galactose-containing surface receptors. Originally it was thought that the mode of action of Ricin poisoning was due to hemagglutination due to a closely related, co-isolating lectin, RCA. |
__NOTOC__ | __NOTOC__ | ||
==Mechanism of action== | ==Mechanism of action== | ||
| - | The mechanism deployed by Ricin to gain entry to a host cell involves the poison's heterogenic properties. First, the B subunit binds to two carbohydrates on the cell surface, either glycolipids or glycoproteins, which both terminate with galactose. The interaction is facilitated by hydrogen bonds to <scene name='Sandbox_BCMB402_Ricin/B_chain_bind_lactose_1/2'>lysine 40 and asparagine 46</scene> in one domain and <scene name='Sandbox_BCMB402_Ricin/B_chain_bind_lactose_2/1'>asparagine 255</scene> in the other domain. Once bound, the ricin-glycoprotein complex is taken into the cells via endocytosis, and the toxin is released into the cells. This association between the A and B chain is essential for toxicity<ref name="montfort" /> without it the Ricin would not be able to gain access to the | + | The mechanism deployed by Ricin to gain entry to a host cell involves the poison's heterogenic properties. First, the B subunit binds to two carbohydrates on the cell surface, either glycolipids or glycoproteins, which both terminate with galactose. The interaction is facilitated by hydrogen bonds to <scene name='Sandbox_BCMB402_Ricin/B_chain_bind_lactose_1/2'>lysine 40 and asparagine 46</scene> in one domain<ref name = "Rutenber">PMID: 3561502</ref> and <scene name='Sandbox_BCMB402_Ricin/B_chain_bind_lactose_2/1'>asparagine 255</scene> in the other domain. Once bound, the ricin-glycoprotein complex is taken into the cells via endocytosis, and the toxin is released into the cells. This association between the A and B chain is essential for toxicity <ref name="montfort" /> without it the Ricin would not be able to gain access to the cell, rendering it useless<ref name = "rapak">PMID: 9108055</ref>.. |
Once the A chain gains entry into the cytosol its mechanism for attack of the [[Ribosome|ribosome]] is depurination of a single adenosine residue in a highly conserved portion within the large RNA of the cytoplasmic [[Large Ribosomal Subunit of Haloarcula|large ribosomal subunit]]<ref name="rapak" /> of eukaryotes; in human, the large cytoplasmic ribosomal RNA is called the 28S ribosomal RNA because of its sedimentation properties during ultracentrifugation. The nucleotide depurinated is located within a specific, conserved loop referred to as the <nowiki>'</nowiki>sarcin-ricin loop<nowiki>'</nowiki>; the loop is critical for binding [[elongation factors|elongation factors]] during [[Translation|translation]] of messenger RNA to protein <ref name="holmbergnygard">PMID: 8648651</ref>. Depurination of the single adenosine nucleotide by the toxin results in the inhibition of protein synthesis. | Once the A chain gains entry into the cytosol its mechanism for attack of the [[Ribosome|ribosome]] is depurination of a single adenosine residue in a highly conserved portion within the large RNA of the cytoplasmic [[Large Ribosomal Subunit of Haloarcula|large ribosomal subunit]]<ref name="rapak" /> of eukaryotes; in human, the large cytoplasmic ribosomal RNA is called the 28S ribosomal RNA because of its sedimentation properties during ultracentrifugation. The nucleotide depurinated is located within a specific, conserved loop referred to as the <nowiki>'</nowiki>sarcin-ricin loop<nowiki>'</nowiki>; the loop is critical for binding [[elongation factors|elongation factors]] during [[Translation|translation]] of messenger RNA to protein <ref name="holmbergnygard">PMID: 8648651</ref>. Depurination of the single adenosine nucleotide by the toxin results in the inhibition of protein synthesis. | ||
Revision as of 18:22, 24 April 2013
Ricin is a potent cytotoxin that is synthesized in the endosperm cells of maturing seeds of the castor oil plant (Ricinus communis)[1]. Ricin belongs to a small multi-gene family[2] that is composed of eight members. Ricin is classified as a type II heterodimeric Ribosome Inactivating Protein[1] or RIPs. For toxins in Proteopedia see Toxins.
| |||||||||||
Potential Applications
Updated April 2013
Ricin A chain (RTA)
1j1m, 1ift, 2aai, 1rtc – RTA
3lc9, 3mk9, 2vc4, 1uq4, 1uq5, 1obs, 3bjg, 3srp – RTA (mutant)
Ricin A chain binary complexes
3px8 – RTA preproricin + 7-carboxy-pterin
1br5, 1br6 - RTA + pterin derivative
3px9 - RTA preproricin + furanylmethyl-carbamoyl-pterin
3lc9, 3mk9, 2vc4, 1uq4, 1uq5, 1obs – RTA (mutant)
3hio – RTA + tetranucleotide
3ej5, 1il5 – RTA pyrimidine derivative
2p8n, 1ifs – RTA + adenine
2pjo, 2r2x – RTA + urea derivative
2r3d – RTA + acetamide
2vc3 - RTA (mutant) + acetate
1il3, 1il4, 1il9 – RTA + guanine derivative
1ifu, 1fmp – RTA + formycin
1obt - RTA (mutant) + AMP
1apg – RTA + RNA
3px8 – RTA + formycin monophosphate
Ricin B chain (RTB)
3nbc, 3nbd – CnRTB + lactose – Clitocybe nebularis
3nbe – CnRTB + lactose derivative
3phz – RTB + glycoside – Polyporus squamosus
Ricin A+B chains
2aai - RTA + RTB
3rtj - RTA + RTB + dinucleotide
See Also
References
- ↑ 1.0 1.1 1.2 1.3 Lord JM, Roberts LM, Robertus JD. Ricin: structure, mode of action, and some current applications. FASEB J. 1994 Feb;8(2):201-8. PMID:8119491
- ↑ 2.0 2.1 2.2 Montfort W, Villafranca JE, Monzingo AF, Ernst SR, Katzin B, Rutenber E, Xuong NH, Hamlin R, Robertus JD. The three-dimensional structure of ricin at 2.8 A. J Biol Chem. 1987 Apr 15;262(11):5398-403. PMID:3558397
- ↑ Weston SA, Tucker AD, Thatcher DR, Derbyshire DJ, Pauptit RA. X-ray structure of recombinant ricin A-chain at 1.8 A resolution. J Mol Biol. 1994 Dec 9;244(4):410-22. PMID:7990130 doi:http://dx.doi.org/10.1006/jmbi.1994.1739
- ↑ Rutenber E, Ready M, Robertus JD. Structure and evolution of ricin B chain. Nature. 1987 Apr 9-15;326(6113):624-6. PMID:3561502 doi:http://dx.doi.org/10.1038/326624a0
- ↑ 5.0 5.1 Rapak A, Falnes PO, Olsnes S. Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc Natl Acad Sci U S A. 1997 Apr 15;94(8):3783-8. PMID:9108055
- ↑ Holmberg L, Nygard O. Depurination of A4256 in 28 S rRNA by the ribosome-inactivating proteins from barley and ricin results in different ribosome conformations. J Mol Biol. 1996 May 31;259(1):81-94. PMID:8648651 doi:10.1006/jmbi.1996.0303
_