Sandbox Reserved 592
From Proteopedia
| Line 3: | Line 3: | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
== Background == | == Background == | ||
| - | {| align= | + | {| align=left |
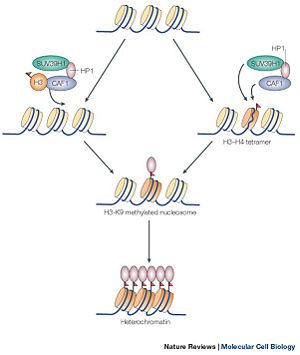
[[Image:picture 1.jpg | 300 px | thumb |Figure 1 showing the methylation of the histone protein and the subsequent formation of heterochromatin]] | [[Image:picture 1.jpg | 300 px | thumb |Figure 1 showing the methylation of the histone protein and the subsequent formation of heterochromatin]] | ||
| Line 51: | Line 51: | ||
== Clinical relevence == | == Clinical relevence == | ||
| - | {| align= | + | {| align=middle |
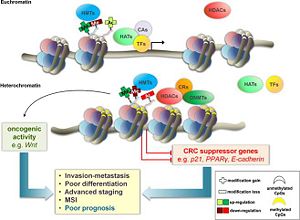
[[Image:picture 3.jpg | 300 px | thumb |Figure 3 showing the methylation of the histone protein and the subsequent formation of heterochromatin]] | [[Image:picture 3.jpg | 300 px | thumb |Figure 3 showing the methylation of the histone protein and the subsequent formation of heterochromatin]] | ||
The physiological state of a cell depends on the physiological state of the genes which defines the function of the cell. Unregulated expression of genes in the cell leads to physiological and morphological change of the cell. Euchromatin and heterochromatin define the physiological state of a cell. Mutations and knockout studies of SUV39H1 in mice have shown to increase genomic instability. The instability arises from the inability of SUV39H1 to form heterochromatin. Since the enzyme could not methylate the nucleosomes and transform a euchromatin state into a heterochromatin state, certain genes were highly expressed, which lead to the instability of cells <ref>PMID:22583735</ref>. | The physiological state of a cell depends on the physiological state of the genes which defines the function of the cell. Unregulated expression of genes in the cell leads to physiological and morphological change of the cell. Euchromatin and heterochromatin define the physiological state of a cell. Mutations and knockout studies of SUV39H1 in mice have shown to increase genomic instability. The instability arises from the inability of SUV39H1 to form heterochromatin. Since the enzyme could not methylate the nucleosomes and transform a euchromatin state into a heterochromatin state, certain genes were highly expressed, which lead to the instability of cells <ref>PMID:22583735</ref>. | ||
Revision as of 08:41, 26 April 2013
==Your Heading Here (maybe something like 'Structure'-- PLEASE DO NOT DELETE THIS TEMPLATE -->
| This Sandbox is Reserved from Feb 1, 2013, through May 10, 2013 for use in the course "Biochemistry" taught by Irma Santoro at the Reinhardt University. This reservation includes Sandbox Reserved 591 through Sandbox Reserved 599. |
To get started:
More help: Help:Editing |
Contents |
Background
SET Structure
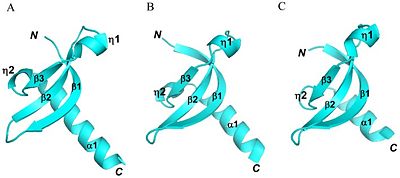
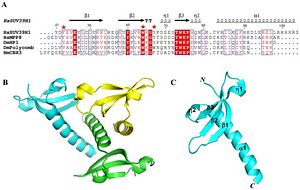
The catalytic domain is consisted of an alpha double helix. The double helix is located at the C-terminus end of the protein. The residue consisting the catalytic domain is about 82-100 amino acids long (Shown in Figure 2). The C-terminus is where the transfer of the methyl group to the lysine residue occurs. In addition to the two main domains to the enzyme, there is an essential hydrophobic core which is very similar to other chromodomain proteins. The hydrophobic core is made up several residues. These residues are V45, L48, Y60, V62, W64, L80, I85 and L86 (Shown in Figure 2). The groove formed by the beta sheets is also similar and conserved feature of the chromodomain family. The physical characteristics between the chromodomain of SUV39h1 and chromodomains of other enzymes are very similar. SUV39h1 has been shown to very similar to the chromodomain of MPP8 and HP1, showing a conservation in chromodomain structure (shown in Figure 3). Although the chromodomain structure is very similar, there is a slight difference with the catalytic domain being longer. In addition to the catalytic domain of SUV39H1 being longer, the enzyme contains a F34 aromatic cage, which was originally thought to be essential for recognizing exposed lysine or argon residue. However, crystallography of residues (44-106) has shown residues missing the F34 aromatic cage.
|