Sandbox Reserved 786
From Proteopedia
(→Overview of Crystalline Structure) |
|||
| Line 17: | Line 17: | ||
| - | The <scene name='56/563198/Unedited/1'> | + | The <scene name='56/563198/Unedited/1'>crystalline structure of malate dehydrogenase</scene> shows that it is a homo-tetramer. However the <scene name='56/563198/Green_helices_red_beta-sheets/2'>biological unit</scene> is a dimer. In the image, the alpha-helices are green, Beta-sheets are red, and random coils (turns) are grey. |
| - | + | ||
== '''Secondary Structure''' == | == '''Secondary Structure''' == | ||
Revision as of 16:35, 20 October 2013
| This Sandbox is Reserved from Oct 10, 2013, through May 20, 2014 for use in the course "CHEM 410 Biochemistry 1 and 2" taught by Hanna Tims at the Messiah College. This reservation includes Sandbox Reserved 780 through Sandbox Reserved 807. |
To get started:
More help: Help:Editing |
|
Contents |
Overview of Mitochondrial Malate Dehydrogenase
Malate dehydrogenase is an enzyme in the citric acid cycle. Overall it is classified as EC 1.1.1.37. Breaking down the classification shows precisely what the enzyme is responsible for doing. "EC 1" generally terms the enzyme as an oxidoreductase, a class of 7,911 protein database entries. An oxidoreductase is an enzyme that is responsible for catalyzing oxidoreduction actions. "EC 1.1" narrows the classification of malate dehydrogenase to an oxidoreductase that is responsible for acting on the CH-OH groups of substrates as donors, a collection of 1,754 protein database entries. "EC 1.1.1" furthers the specificity of the protein to be an oxidoreductase that acts on CH-OH groups using NAD(+) or NADP(+) as acceptors, a group of 1,585 entries in the protein database. The final classification"EC 1.1.1. 37" yields 60 protein database entries, and narrows the definition with the name of the enzyme,malate dehydrogenase, and its specific reaction.
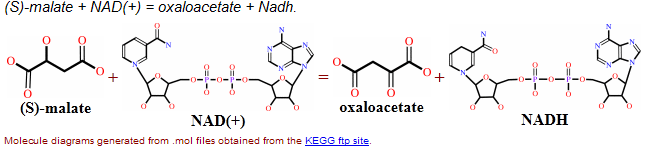
Enzymatic Reaction
Overview of Crystalline Structure
The shows that it is a homo-tetramer. However the is a dimer. In the image, the alpha-helices are green, Beta-sheets are red, and random coils (turns) are grey.
Secondary Structure
The is formed by hydrogen bonding interactions of the amino acids backbone. There are two beta-sheet regions in each monomer of the natural dimer. Five beta-sheets in each monomer are anti-parallel, and form a pseudo beta-barrel motif. Six beta-sheets in each monomer show parallel configuration. The angle of the hydrogen bonds relative to one another between beta-sheets gives insight as to which beta-sheets are parallel and which are anti-parallel. Parallel beta-sheets have "crooked" h-bonding. This makes them more unstable than the anti-parallel beta-sheets which have straight h-bonds between them.