Sandbox Reserved 768
From Proteopedia
| Line 28: | Line 28: | ||
== Phenylalanine Hydroxylase Structure == | == Phenylalanine Hydroxylase Structure == | ||
| - | Although the full-length structure of mammalian PAH has not yet been clearly known, a large part of it has been identified. <ref name= "flydal"/>. It was solved by means of crystallizing the protein (at pH=7) to perform X-ray | + | Although the full-length structure of mammalian PAH has not yet been clearly known, a large part of it has been identified. <ref name= "flydal"/>. It was solved by means of crystallizing the protein (at pH=7) to perform X-ray diffraction using molecular replacement. The search model used was based on the crystal structure of tyrosine hydroxylase because of the similarity between the two enzymes. <ref name="fusetti"> Fusetti, Fabrizia, Heidi Erlandsen, Torgeir Flatmark, and Raymond Stevens. "Structure of Tetrameric Human Phenylalanine Hydroxylase and Its Implications for Phenylketonuria." The Journal of Biological Chemistry 273.27 (1998): 16962-16967. Web. </ref>. The R-factor recorded was 0.251 and the mean B (or temperature) value was 33.0. <ref name= "pdb"/>. |
'''Structure Revealed''' | '''Structure Revealed''' | ||
| Line 44: | Line 44: | ||
The C-terminal oligomerization or tetramerization domain begins with an antiparallel-sheet | The C-terminal oligomerization or tetramerization domain begins with an antiparallel-sheet | ||
(residues 411–414, 421–424) | (residues 411–414, 421–424) | ||
| - | and is formed by a C-terminal “arm” consisting of two β-strands, forming a β-ribbon, and a 40 Å long α-helix. This C-terminal arm extends over an adjacent monomer, thus bringing the four helices (one from each monomer) into a closely packed anti-parallel coiled-coil motif in the center of the structure (as can be seen in the tetramer structure above). <ref name= "fusetti"/>. | + | and is formed by a C-terminal “arm” consisting of two β-strands, forming a β-ribbon, and a 40 Å long α-helix. This C-terminal arm extends over an adjacent monomer, thus bringing the four helices (one from each monomer) into a closely packed anti-parallel coiled-coil motif in the center of the structure (as can be seen in the tetramer structure above). <ref name= "fusetti"/>. The assembly of the enzyme occurs through a swapping mechanism in which the secondary structural elements mutually switch their position to promote oligomerization. <ref name="fusetti"/>. |
Revision as of 02:41, 2 December 2013
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
|
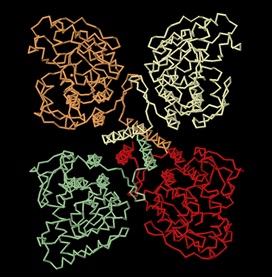
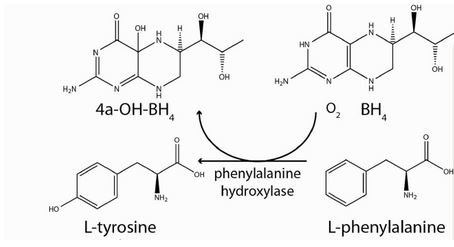
Phenylalanine Hydroxylase (also known as Phenylalanine-4-monooxygenase or simply PAH) is the enzyme that catalyzes the conversion of L-phenylalanine into L-tyrosine by hydroxylation (addition of an -OH group) of the aromatic side chain of phenyalanine. This reaction is the initial and rate-limiting step in the phenylalanine catabolism pathway. The tyrosine product (a non-essential amino acid) can then serve as a precursor to the synthesis of important neurotransmitters.[1]. PAH uses tetrahydrobiopterin (BH4)as a cofactor and has a nonheme iron atom bound to its active site. PAH is classified as an oxidoreductase, specifically enzyme class EC 1.14 since its mechanism of action involves the oxidation/reduction of its substrate. [2]. PAH enzyme is present in the liver cells of humans and other mammals. It is also present in non-mammalian eukaryote organisms and some bacteria such as E.coli. [2]. Recently, it has been identified in some protozoans and slime molds, and even in nonflowering plants such as spinach from which it has been extracted and studied. [3]. Mammalian PAH is a homo-tetrameric enzyme of 50 kDa subunits composed of two asymmetric dimeric units. The four subunits are connected to each other via a coiled-coil motif as shown in the diagram below.
Each monomeric subunit is composed of three sites: the N-terminal, the catalytic site, and the C-terminal. [1]. PAH enzyme has been studied extensively because of its correlation with the genetic defective condition phenylketonuria (PKU). Errors in the function or stability of PAH lead to its malfunction which causes a buildup of phenylalanine resulting in numerous health detriments.
Phenylalanine Hydroxylase Mechanism of Action
PAH, belonging to the oxidoreductase enzyme class, acts by oxidizing/reducing its substrate. Specifically, PAH acts on paired donors with pteridine being a donor and first incorporates one atom of molecular oxygen into the aromatic ring of phenylalanine. [2]. Then, it reduces the second oxygen atom to water using the two electrons that are supplied by the BH4 cofactor. BH4 is also hydroxylated at each turnover to produce pterin-4a-carbinolamine (4a-OH-BH4), with consequent dissociation from the enzyme. 4a-OHBH4 is dehydrated and reduced back to BH4 by the action of the enzyme pterin carbinolamine dehydratase. [1].
Kinetics Overview
Formation of the productive PAH−BH4−phenylalanine complex begins with the rapid binding of BH4 (Kd = 65 μM). Subsequently, phenylalanine is added to the binary complex to form the productive ternary complex (Kd = 130 μM). This second step is approximately 10-fold slower. Both substrates are able to bind to the free enzyme form to produce inhibitory binary complexes. Molecular oxygen rapidly binds to the formed productive ternary complex; which is then followed by formation of an unidentified intermediate. This intermediate can be detected as a decrease in absorbance at 340 nm, with a rate constant of 140 s−1. Formation of the 4a-OHBH4 and Fe(IV)O intermediates is 10-fold slower and is followed by the rapid hydroxylation of the amino acid. Product release is the rate-determining step and largely determines kcat. [4].
Phenylalanine Hydroxylase Structure
Although the full-length structure of mammalian PAH has not yet been clearly known, a large part of it has been identified. [1]. It was solved by means of crystallizing the protein (at pH=7) to perform X-ray diffraction using molecular replacement. The search model used was based on the crystal structure of tyrosine hydroxylase because of the similarity between the two enzymes. [5]. The R-factor recorded was 0.251 and the mean B (or temperature) value was 33.0. [2].
Structure Revealed
The monomeric unit of PAH is composed of three sites: an N-terminal regulatory domain (residues 1–110), a core catalytic domain (residues 111–410), and a C-terminal oligomerization or tetramerization domain (residues 411–452). [1].
The N-terminal regulatory domain is a regulatory module present in several proteins, which functions in the dimerization and binding of amino acids. It is flexibly attached to the catalytic domain via a hinge region (Arg111–Thr117). [1].
The catalytic domain contains the active site of the enzyme. It is composed of 13 α-helices and 8 β-strands [5] and houses the binding sites for the nonheme iron atome, the cofactor, and substrate. The iron binds to two histidines (His285 and His290 in hPAH) and a glutamate (Glu330) in the deep cleft in the core of each monomer. [6].
The C-terminal oligomerization or tetramerization domain begins with an antiparallel-sheet (residues 411–414, 421–424) and is formed by a C-terminal “arm” consisting of two β-strands, forming a β-ribbon, and a 40 Å long α-helix. This C-terminal arm extends over an adjacent monomer, thus bringing the four helices (one from each monomer) into a closely packed anti-parallel coiled-coil motif in the center of the structure (as can be seen in the tetramer structure above). [5]. The assembly of the enzyme occurs through a swapping mechanism in which the secondary structural elements mutually switch their position to promote oligomerization. [5].