Sandbox Reserved 760
From Proteopedia
| Line 12: | Line 12: | ||
The first method for crystallization of nitrogenase was put forward by Burns R.C., Holsten R.D. and Hardy R.W., then was modified by Shah V.K. and Brill W.J. and was simplified by the 7th Laboratory, Institute of Botany, The Chinese Academy of Sciences. But the crystals gotten from these laboratories were small and needle-shaped therefore wasn’t suitable for X-ray diffraction analysis. Weininger M.S. and Mortenson L. E. attempted to do X-ray diffraction analysis of MoFe protein crystals grown by using microdialysis method but they did not succeed in analyzing the crystallographic structure of MoFe protein. Later Kim and Rees succeeded in growing the big crystals of Azotobacter vinelandii nitrogenase by the microcapillary batch method and in analyzing of these crystals by X-ray diffraction. Since then, the liquid/liquid diffusion method has been usually used for crystal growth of nitrogenase proteins. | The first method for crystallization of nitrogenase was put forward by Burns R.C., Holsten R.D. and Hardy R.W., then was modified by Shah V.K. and Brill W.J. and was simplified by the 7th Laboratory, Institute of Botany, The Chinese Academy of Sciences. But the crystals gotten from these laboratories were small and needle-shaped therefore wasn’t suitable for X-ray diffraction analysis. Weininger M.S. and Mortenson L. E. attempted to do X-ray diffraction analysis of MoFe protein crystals grown by using microdialysis method but they did not succeed in analyzing the crystallographic structure of MoFe protein. Later Kim and Rees succeeded in growing the big crystals of Azotobacter vinelandii nitrogenase by the microcapillary batch method and in analyzing of these crystals by X-ray diffraction. Since then, the liquid/liquid diffusion method has been usually used for crystal growth of nitrogenase proteins. | ||
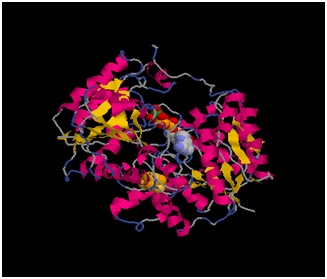
| - | [[Image:tdd.jpg]] | + | [[Image:tdd.jpg]]Fe-Protein |
| + | The Fe-protein is a 60 kilo dalton dimer of identical subunits with a single cluster of Fe4S4 in between the two subunits. The three-dimensional crystal structure of Fe-protein symmetrically coordinated from each subunit. Significantly, the subunits have the α-helical and β-sheet type of shape commonly associated with a major class of nucleotide-binding proteins. The two subunits are related by a molecular two-fold rotation axis that passes through the iron-sulfur cluster, which is located at the center of the dimer, right at the interface of the subunits.The iron-sulfur cluster not only serves as a crosslink between both subunits but it helps stabilize the dimer structure along with some salt and hydrophobic interactions. The Fe protein has two main functions; one: binding MgATP, MgADP, and nucleotides. Two: the second functional site in the Fe protein is the Fe4S4 cluster, which undergoes a one electron redox cycle between the 2Fe2+2FeH state and the 3Fe2+FH state. Both cluster ligands are located near the amino-terminal end of the helices that are directed towards the cluster. NH-S bonds are form to provide stabilizing electrostatic forces to the center of the protein. Other than the cysteinyl ligands, there is little contact between the Fe4S4 cluster and other amono acid side chains. The image of the Fe protein cluster is that of an exposed and loosely packed center that acts as a pivot or hinge to allow any conformational change between subunits during the reaction of the enzyme. When the subunits pivot or swing together it is lowered towards the FeMo protein to ensure electron transfer as shown in the diagram below. | ||
| + | |||
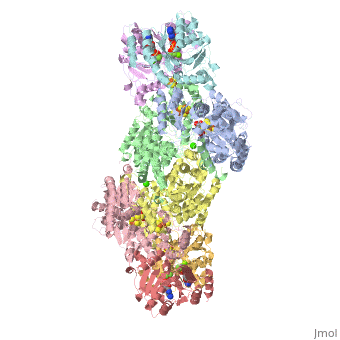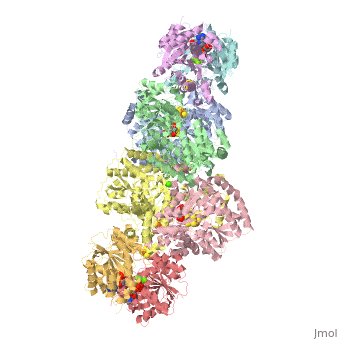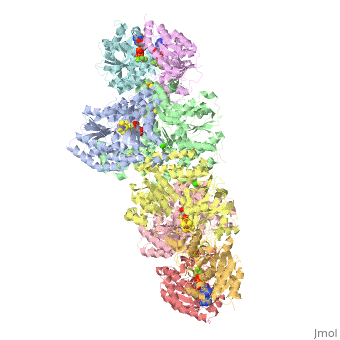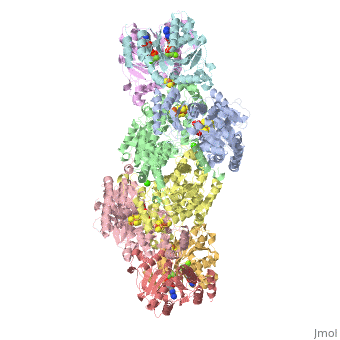
<Structure load='1N2C' size='500' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | <Structure load='1N2C' size='500' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | ||
Revision as of 08:16, 6 December 2013
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
INTRODUCTION What comes to the mind when one thinks about nitrogenase is the nitrogen cycle in which nitrogen is converted from its naturally occurring form to ammonia. Biological nitrogen fixation, the reduction of atmospheric N2 to NH3 by nitrogen fixing organism, is a key step in the nitrogen cycle. Nitrogenase is an enzyme (protein) that fixes nitrogen (N2) from the atmosphere into ammonia (NH3). This enzyme is only found in prokaryotic organisms such as certain bacteria and blue-green algae in which some are free living (Cyanobacteria) or symbiotic (Rhizobia) in nature.
Nitrogenase catalyzes the following reaction: N2 + 8H+ + 16MgATP + 8e− → 2 NH3 + H2 + 16MgADP + 16Pi.
8 electrons are supplied to the reaction via the reduction of nitrogenase coupled by the presence of twice the amount of ATP and magnesium ions. The substrate (N2) is then reduced in the active site of the enzyme along with 8hydrogen ions to form 2 ammonia molecules and a by-product of hydrogen. The hydrolyzed ATP gives off ADP and inorganic phosphate. This process is catalysed by nitrogenase, the most common form of which is the molybdenum nitrogenase. The molybdenum nitrogenase is made up of dinitrogenase also known as Molybdenum-iron protein (MoFe protein) or NifDK, a double α and double β tetramer encoded by the nifD and nifK genes, respectively. Molybdenum nitrogenase has another protein, dinitrogenase reductase also known as iron protein (Fe protein) or NifH, a γ2 homodimer encoded by the nifH gene. The NifH protein is responsible for ATP-hydrolysis-driven electron transport to the NifDK protein, which contains the site for the reduction of dinitrogen to ammonium. The reduction of N2 to two molecules of NH3 is an energy consuming process that requires 16ATPs, 2ATPs per electron used in the reaction.
STRUCTURE The first method for crystallization of nitrogenase was put forward by Burns R.C., Holsten R.D. and Hardy R.W., then was modified by Shah V.K. and Brill W.J. and was simplified by the 7th Laboratory, Institute of Botany, The Chinese Academy of Sciences. But the crystals gotten from these laboratories were small and needle-shaped therefore wasn’t suitable for X-ray diffraction analysis. Weininger M.S. and Mortenson L. E. attempted to do X-ray diffraction analysis of MoFe protein crystals grown by using microdialysis method but they did not succeed in analyzing the crystallographic structure of MoFe protein. Later Kim and Rees succeeded in growing the big crystals of Azotobacter vinelandii nitrogenase by the microcapillary batch method and in analyzing of these crystals by X-ray diffraction. Since then, the liquid/liquid diffusion method has been usually used for crystal growth of nitrogenase proteins.
The Fe-protein is a 60 kilo dalton dimer of identical subunits with a single cluster of Fe4S4 in between the two subunits. The three-dimensional crystal structure of Fe-protein symmetrically coordinated from each subunit. Significantly, the subunits have the α-helical and β-sheet type of shape commonly associated with a major class of nucleotide-binding proteins. The two subunits are related by a molecular two-fold rotation axis that passes through the iron-sulfur cluster, which is located at the center of the dimer, right at the interface of the subunits.The iron-sulfur cluster not only serves as a crosslink between both subunits but it helps stabilize the dimer structure along with some salt and hydrophobic interactions. The Fe protein has two main functions; one: binding MgATP, MgADP, and nucleotides. Two: the second functional site in the Fe protein is the Fe4S4 cluster, which undergoes a one electron redox cycle between the 2Fe2+2FeH state and the 3Fe2+FH state. Both cluster ligands are located near the amino-terminal end of the helices that are directed towards the cluster. NH-S bonds are form to provide stabilizing electrostatic forces to the center of the protein. Other than the cysteinyl ligands, there is little contact between the Fe4S4 cluster and other amono acid side chains. The image of the Fe protein cluster is that of an exposed and loosely packed center that acts as a pivot or hinge to allow any conformational change between subunits during the reaction of the enzyme. When the subunits pivot or swing together it is lowered towards the FeMo protein to ensure electron transfer as shown in the diagram below.
|