| 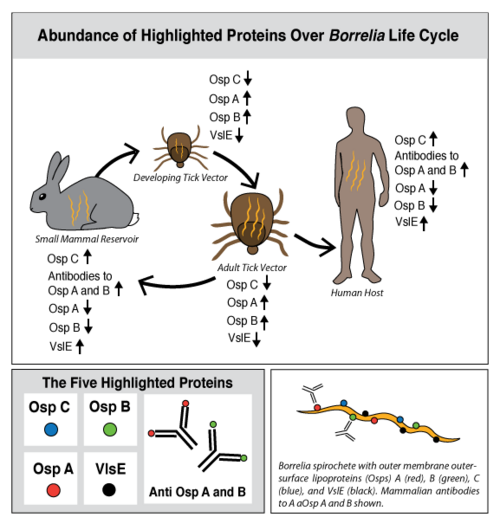 Figure 1: Abundance of Highlighted Proteins Over Borrelia Life Cycle. Lyme disease is caused by three species of bacteria belonging to the genus Borrelia.[1][2] Borrelia burgdorferi, an obligate parasite, is the most common cause of the disease in the United States and is transmitted via hard-bodied ticks of the Ixodidae family, commonly known as the blacklegged or deer ticks. Borrelia spirochetes are motile, helical bacteria whose cell membranes have many exposed, surface lipoproteins that are involved in both the pathogenesis and life cycle of the parasite. Two predominant groups of the surface lipoproteins present are classified as outer surface proteins (Osps), which have been characterized as Osps A through F, and the variable major protein-like sequence expressed (VlsE). Both of these groups of outer surface proteins play important roles in both the pathogenesis and life cycle of Borrelia as well as roles in eliciting an immune response of the host organism (Figure 1).[3]
</p>
In a introductory biology course at Stony Brook University, undergraduates are modeling and exploring B. burgdorferi outer surface proteins and their respective antibodies in which the host organism produces. This Proteopedia page is the product of their efforts, with a focus on highlighted proteins from the following categories: OspC, OspB and the antibodies to OspB, OspA and the antibodies to OspA, and VlsE.
The goal of this Proteopedia page is to describe Lyme disease from a structural biology perspective in order to answer key questions regarding the relationship between the structure and function of B. burgdorferi proteins. What do B. burgdorferi outer surface proteins look like? How does the structure/function of these proteins relate to the infection cycle of B. burgdorferi? What are the structural targets of the human immune system and how have these targets evolved? What are the ideal structural targets for a vaccine to protect against Lyme disease?
OspC and Lyme Disease
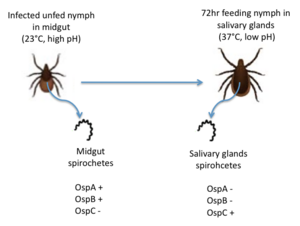 Figure 2: The migration of B. burgdorferi from the midgut to the salivary glands in infected nymphs. Redrawn from Templeton 2004. OspC, a highly variable B. burgdorferi outer surface protein, plays a pivotal role in the transmission of B. burgdorferi from the tick vector to a mammalian host. This protein is upregulated when the tick feeds, allowing for it to adhere to the tick saliva, move to the tick's mouth, and migrate into the mammalian host.[4] The upregulation of OspC is accompanied by a downregulation of two other outer surface proteins, OspA and OspB, which is thought to be induced by changes in environmental temperature and pH (Figure 2). [5]
Strains of B. burgdorferi are classified according to the sequence of the OspC locus into 19 outer surface major groups (oMGs), denoted by type A through S, of which only four are invasive (disease causing).[6] Polymorphisms of OspC and the abundance of invasive strains in a population of B. burgdorferi are driven by ecological factors, such as the mammalian host community composition, and are determinants of human Lyme disease risk[7].
Researchers are attempting to take advantage of the upregulation of OspC on B. burgdorferi's outer surface while the bacteria is in the host to develop an OspC-based vaccine. However, development of an OspC-based vaccination has been presented with difficulties due to its highly variable nature. [8]
Structure of OspC
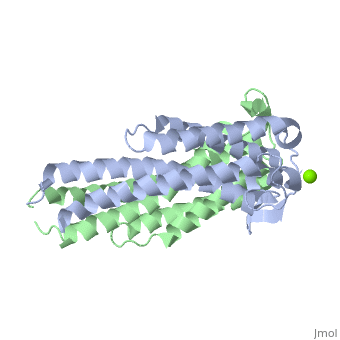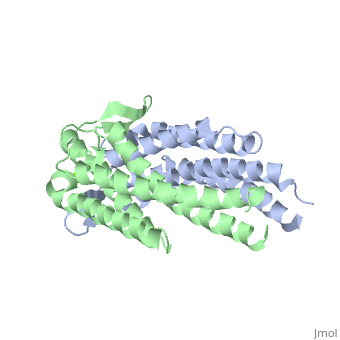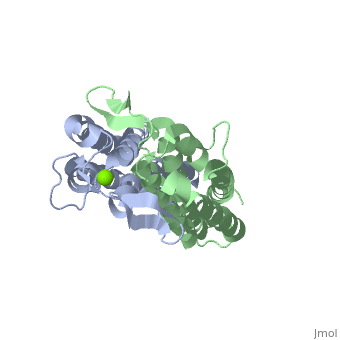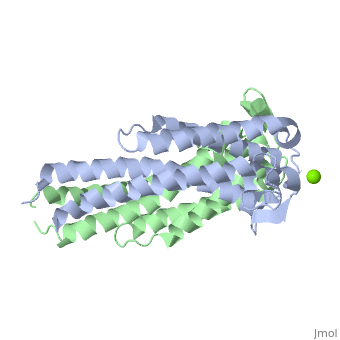
The OspC three-dimensional model presented to the right is the B. burgdorferi B31 strain (residues 38-201) - also known as the oMG A strain. This is one of the four invasive oMGs that are responsible for systematic Lyme disease in mammalian hosts. In crystal structure, OspC exists as a dimer that coordinates a divalent ion, modeled here as magnesium. Each OspC subunit is predominantly helical, consisting of five parallel
, two short, antiparallel
and six
. The N and C termini at the membrane proximal end of two long α-helices,
(residues 38-76) and
(residues 170-201), are in close proximity to each other. At the membrane distal end, there are three remaining alpha helices,
(residues 95-112),
(residues 121-145), and a the final, short
(residues 152-159). At the end of the membrane surface, the connection between helices α1 and α2 forms two short, anti-parallel, β-strands,
(residues 79-80),and
(residues 88-89).
While most of the OspC locus is highly variable, the sequence alignment of all oMGs reveals that the surface-exposed residues towards the membrane-proximal end of two of the helices, α1 and α5, are highly and have a positively charged surface. Other than those regions of the α1 and α5 helices, the surface-exposed residues on the remaining regions of the OspC molecule are variable.[9]
OspC Structure and Antigenicity
At the membrane-distal region, the six loop regions, including two β-strands, illustrates the
of OspC with the presence of variable surface-exposed residues. [10] Among these variable regions, the outer surface-exposed residues connecting the helices α1 and α2, forming the loops,
(residues 74-78),
(residues 81-87),
(residues 90-93), two short beta strands, β1 and β2, and
(residues 146-150)
are more variable than those present in the loops,
(residues 115-119)and
(residues 161-169). The surface potential of the red region that projects away from the membrane is negatively charged and primarily involved in the protein-protein or protein-ligand interactions.[9] Only four types of invasive oMG strains (A, B, I and K), whose surface potential in the red region is highly negative relative to non-invasive strains, play a major role in pathogenesis of human Lyme disease. [6]The residue, located in the red region at the distal, membrane end, is unique in that replacing this residue with other residues, with the exception of Lys82 and Gln82, which are only present in four invasive oMG strains, enhances the possibility of turning invasive strains into non-invasive strains. Thus, the stronger the negative electrostatic potential in the red region, the higher the chance OspC will to bind with positively charged host ligands; therefore, the altering of an amino acid residue at the 82nd position in the red region determines OspC polymorphism and demonstrates how this is connected to virulence and invasiveness.[6]
Lyme Disease and Ecology
![Figure 3: A Diagram of the Life Cycle of the Blacklegged Tick.[[1]]](/wiki/images/thumb/7/7d/Life_cycle_of_tick.png/300px-Life_cycle_of_tick.png) Figure 3: A Diagram of the Life Cycle of the Blacklegged Tick.[ [1]] Ticks are born with the absence of the B. burgdorferi parasites and acquire them while feeding on the blood of natural, reservoir hosts, such as mice, squirrels, shrews, and other small vertebrates (Figure 3). After growth and development, the infected nymphal and adult ticks can transmit B. burgdorferi to incidental vertebrates, including humans. The ecological interaction between the competence of reservoir hosts and the ticks is an underlying measure of human Lyme disease risk.[11] The occurrence of Lyme disease is dependent upon the abundance of ticks that are infected with B. burgdorferi in natural ecosystems, and the number of reported cases of Lyme disease continues to increase annually in highly focused geographic locations of the United States (Figure 4).
Using Ecological Models to Predict Lyme Disease Risk
![Figure 4: Illustrated Prevalence of Lyme Disease in the United States (Generated by the CDC).[[2]]](/wiki/images/thumb/9/9d/Lyme_Disease_Risk_Map.gif/300px-Lyme_Disease_Risk_Map.gif) Figure 4: Illustrated Prevalence of Lyme Disease in the United States (Generated by the CDC).[ [2]] Researchers have developed conceptual and mathematical models to characterize the ecological interactions between the community of vertebrate hosts and distribution frequency of invasive oMGs and predict the distribution of Lyme disease. In one effective model, the principal reservoir host used to calculate the risk of Lyme disease in northeastern and central United States is of the white-footed mouse (Peromyscus leucopus). The white-footed mouse has both a high frequency distribution in all four human infectious oMGs and high transmission probabilities of oMGs A, B, I and K.[7] It has been found that decreasing the abundance of mammalian hosts with high transmission probabilities, such as the white-footed mouse, and increasing other mammals with lower transmission probabilities drastically decreases the human Lyme disease risk. Many studies have found support for this "dilution-effect model," indicating that maintaining a high diversity of the vertebrate host community may be helpful in decreasing the incidence of Lyme disease. This model strongly demonstrates the relationship between species diversity in the community of hosts and the risk of human exposure to Lyme disease, and the ecological driving forces described in the model are useful tools in predicting the prevalence and risk of human Lyme disease.
Ecological factors responsible for human Lyme disease risk
The following are some factors used as parameters in ecological Lyme disease models:
- Vertebrate Community Composition[12]: Both the abundance and diversity of the mammalian hosts living in the community strongly affects the proportion of infected nymphal ticks that can cause human Lyme disease.
- Distribution frequency of particular oMGs[7]: As only four types of oMGs (A, B, I and K) are responsible for systemic human Lyme disease, the host-seeking nymphs that have a high distribution frequency of the four invasive oMGs is one of the standard measures of human Lyme disease risk.
- Transmission Probability[7]: The transmission probability of each oMG between individual species differs. The higher the transmission probability of a particular oMG from a vertebrate host, the higher the chances are that the ticks will carry that particular oMG after receiving a blood meal from their hosts. Thus, it is one of the parameters that contributes to the prevalence of human Lyme disease.
OspC-based vaccine
Researchers are currently developing an OspC-based vaccine against human Lyme disease, but because of the variability of OspC, the recombinant OspC vaccine, targeting the antigenic site of one B. burgdorferi strain, may be ineffective for other strains. Therefore, the development of a vaccine that recognizes the antigenic determinant on the variable regions of multiple OspC strains is required in order to effectively activate the human immune response. Based on the mapping of epitope-containing regions from oMGs A, B, K and D, the experiment-based tetravalent chimeric vaccine is being developed to trigger an anti-ABKD response. [13] Taking advantage of the tetravalent ABKD construct, an octavalent chimeric vaccine, also known as OspC-A8.1, recognizing additional epitopes of oMGs C, E, N and K has been tested in mice. [8]
OspB and Lyme Disease
OspB and OspA comprise the most common proteins found on the surface of B. burgdorferi while residing in the tick vector, during late-stage Lyme disease, and also during culture conditions.[14] OspB has been found to play a vital role in the adherence of B. bugdorferi to the tick midgut whereas OspB-deficient B. burgdorferi are shown to bind poorly to tick gut extracts. While the tick remains unfed, the expression of both OspB and OspA is upregulated to promote binding to the tick’s gut. However, during transmission from the tick to a vertebrate host, OspB is downregulated while other proteins, such as OspC, are upregulated [15]. OspB has shown significant variability in amino acid sequence and antigen reactivity in comparison to OspA, which is known to be largely invariant [16].
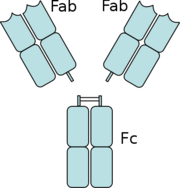 Figure 5: The separation of the Fab domains from the Fc domains of an antibody by Papain digestion.
A factor contributing to the severity of Lyme disease is the ability of B. burgdorferi to evade the host immune system. B. burgdorferi is an extracellular pathogen, which is targeted by the humoral immune system of the host; the complement system and antibodies produced by B-lymphocyte-derived plasma cells work together in order to fight off the infection. The complement system consists of three pathways: the classical pathway, the alternative pathway, and the lectin pathway. Although each pathway differs in the process of initiation, each pathway results in the amplification of an immune response and the formation of the membrane attack complex (MAC), which ultimately leads to the pathogen's demise. In the lectin pathway and the alternative complement pathway, complement is recruited to the pathogen surface directly, but in the classical complement pathway, recruitment is dependent on the existence of an antibody-antigen immune complex; therefore, the traditional view of antibodies functioning in a bactericidal capacity has required complement recruitment and MAC formation following antibody binding to pathogens. Generally, antibodies are thought to be non-bactericidal in the absence of complement.
B. burgdorferi has developed resistance to a complement-dependent immune response by the evasion of the classical complement pathway [17][18] and the alternative complement pathway by binding complement components C4b and CS, respectively.[19] However, OspB is an important target of antibodies that can kill the bacteria without the help of the complement system.[3] These antibodies are termed complement-independent bactericidal antibodies. One important complement independent antibody whose bactericidal role has been well researched is CB2.[20] CB2 has been shown to kill bacteria in vitro in the absence of complement [21]. Furthermore, it has been shown that a point mutation in OspB renders the epitope unrecognizable by CB2, preventing CB2 from binding OspB and suggesting a possible mechanism for the evolution of the bacteria to evade this aspect of the immune system[22].
Another complement independent antibody is H6831, an IgG antibody that targets the C-terminal of OspB (Figure 6). Studies on both CB2 and H6831 have been conducted using the Fab (Fragment antigen binding) portion of these antibodies. A Fab consists of a heavy chain and light chain, each chain containing both a variable region as well as a constant region (Figure 5). The complementarity-determining regions (CDRs) are located at the N-terminal end of the variable region of the heavy and light chains of the Fab and form unique tertiary and quaternary protein structures that determine the antigen binding specificity. Binding of this region of the Fab to OspB of B. burgdorferi leads to the lysis of the bacteria, ruling out simple agglutination (clumping) of the bacteria as the cause of the bactericidal effect.[3]
Structure of the OspB-H6831 Complex
| |
: ·· ·· ··
:
·· ··
:
:
</center> | </table>
The consists of two components: outer surface protein B () and the , which is subdivided into the and the . Most hydrogen bonds and electrostatic interactions that are responsible for the binding of H6831 to OspB are between the at the C-terminal of OspB and some that include tyrosine, tryptophan, glutamate, and histidine.[16]
The majority of hydrogen bonds and electrostatic interactions are between (residues 250-254) and the Fab heavy chain. in loop 2 of OspB has a necessary and major role due to its central position in the exposed loops. A mutation at its position abrogates the binding interaction and causes the resistance of the bacteria to the bactericidal effect of the Fab. Lys 253 interacts with the two aromatic residues on the Fab heavy chain, tyrosine and tryptophan. It also makes hydrogen bonds with the glutamate 50 in the heavy chain of the Fab and forms an ionic bond. Carbonyl in of the OspB interacts with in the Fab heavy chain. of OspB interacts with Fab light chain.[16]
Structural changes to OspB in the complexed form
The binding of H6831 to OspB leads to some conformational changes in OspB compared to its . Crystallography has shown that the most significant difference is the loss of the .[16] The loss of these β sheets may be due to conformational change as a result of the binding or a disorder that could have occurred during a crystallization of the complex. Both small positional shifts near the Fab binding site and a few larger structural changes away from the binding site were observed. The largest shifts (7– 8 Å) correspond to the repositioning of a loop opposite the Fab-binding site . In the free OspB structure, all regions that exhibit shifts are adjacent to the central sheet; in the OspB-H6831 complex they all shift toward, and slightly overlap the position of the missing sheet.
Potential Mechanism of Lysis
The Fab binding destabilizes the outer membrane (OM) of B. burdorferi, with subsequent formation of spheroplasts. It has been observed that the bactericidal action, but not the binding, requires the presence of divalent cations (Mg2+ and Ca2+), and
Fab is unable to clear bacteria in the absence of these cations.[23] It is speculated that OspB-Cb2 (a Fab similar to H6831) complexes could lead to the lysis of the cell by creating physical openings in the OM, allowing for rapid infusion of electrolytes and increasing the osmolarity of the periplasm.[24]
Due to its effective bactericidal actions, H6831 is used to generate less virulent escape variants of B. burgdorferi. [16] In the majority of the mutations created from in vivo and in vitro immunization of mice, truncated forms of OspB within the C terminus lead to premature stop codons.[25] It has been suggested that OspB mutants are more sensitive to proteolysis due to missense mutations that disturb the conformation of OspB [16]. Truncated OspBs cease within the two C-terminal beta-strands of the central sheet. H6831 disorders or removes a beta sheet from OspB after binding. Cleavage may be a possible explanation for the conformational changes of OspB. [24] In and forms of OspB, some changes result from proteolysis near the N terminus [16]. Residues 157 - 201 on OspB contain the , shown in pink.
Aromatic residues tyrosine and tryptophan are also present in the OspB-H6831 interaction, a feature found in many antigen-antibody complexes. The Lys-253 residue forms a trans conformation between these aromatic residues of H6831. In the complex structure of the antibody binding site, the electron density is well defined and shows increased contact between Lys-253 and the antigen-binding site of the Fab. Most of the electrostatic and hydrogen-bond interactions occur between loop 2 and the Fab heavy chain.
Potential Catalytic Triad
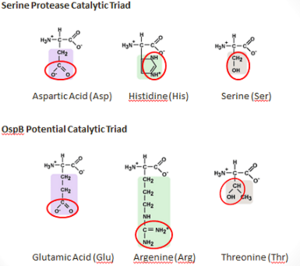 Figure 9: Comparison of the Compositions of the OspB and a Serine Protease Catalytic Triads The mechanism by which H6831 Fab destroys a spirochete appears to be a novel interaction. It is possible that Fab binding changes the properties of OspB folding, which may increase sensitivity of the protein to proteolysis or aggregation. NMR methods showed that the effects of binding can be sent to regions of the antigen distant from epitope, which is at the shown in red (N-terminus in blue). OspB shows signs of truncation after interacting with Fab of H6831 [26].
It is possible that OspB performs an autoproteolysis. There is a found on OspB that resembles the catalytic triad of Serine_Proteases. This "constellation" consists of Thr-166, Arg-162, and Glu-184, which is similar to the catalytic triad residues of the serine protease trypsin, which are Ser-195, His-57, Asp-102 [27].
Threonine and Glutamic acid are found in other catalytic triads of the serine hydrolase family, but argenine seems unlikely to replace histidine as a base because of its higher pKa. There have been studies that have shown that Argenine is essential for other enzymatic functions, such as in the Ser-Arg-Asp triad in cytosolic phospholipase A2 and as a catalytic base in Sortase A. forms an H-bond with and may rearrange to form a putative oxyanion hole with Thr-166 and another unidentified atom if active in the catalysis. A concerted proton transfer, similar to a “proton wire”, is one plausible mechanism that would allow argenine to function in the catalytic triad of a protease.
OspA and Lyme Disease
Like OspC and OspB, OspA's expression is differentially regulated over the B. burgdorferi infection cycle. OspA is expressed while the bacteria resides in the midgut of the tick, downregulated while the tick feeds on its host, and then upregulated in the host's cerebrospinal fluid (CSF), which may induce an inflammatory response resulting in acute Lyme neuroborreliosis. As mentioned above, OspA, together with OspB, make up the major proteins found on the surface of B. burgdorferi. Because OspA is relatively highly expressed and relatively invariable it has been used as a target in the development of a vaccine for Lyme disease.
OspA is involved in attachment of B. burgdorferi to the tick gut by binding to the Tick receptor for OspA (TROSPA). TROSPA is a tick gut protein which is required for colonization of the spirochete[28]. When a tick feeds, OspA is downregulated, releasing the B. burgdorferi from the gut wall and allowing the bacteria to migrate into the tick's salivary glands and then into the host. The downregulation of OspA during transmission is evidenced by the fact that patients with Lyme disease do not possess OspA antibodies in the early stages of the disease.[3][29]
Structure of OspA
| |
|
,
in C-terminus (close up) (residues 203-220) (residues 224-233) (residues 246-257) in C-terminus (Ala208, Ala215 and Asn251 in B. burgdorferi; also hides R-groups). of Ala208, Ala215 and Asn251
|
OspA is made up of 273 residues over 21 anti-parallel β-sheets and a single α-helix. It's folded conformation is divided into three main sections: a N-terminus "sandwich," a central region comprising of several β-sheets and a C-terminus "barrel" domain.[23] The folded regions at its ends are connected by a single β-sheet layer in the middle, giving the protein the unique shape of a dumbell.[30]
There are at the C-terminus of OspA that are important in binding with the LA-2 Fab antibody, whose interactions provide great insight into vaccine research and effectiveness. These three loops are linearly arranged and form protruding ridge at the C-terminus of OspA. Within these loops, there are where there are distinct variations between the different strains of B. burgdorferi and serve as potential targets for the creation of a broader vaccine.[23]
, (residues 203-220), is important in showing variation amongst the different strains of B. burgdorferi as well as being optimally conformed for binding without steric hindrance. (residues 224-233) and (residues 246-257) are more strongly conserved than Loop 1 but also help to show some variation amongst strains. The LA-2 Fab antibody readily recognizes OspA from B. burgdorferi, but does not recognize that from B. afzelii or B. garinii.
Between B. burgdorferi and B. afzelii genetic sequences are generally invariant, but two residues change between the species: in B. burgdorferi is a Glutamine (Gln) in B. afzelii, and in B. burgdorferi is an Alanine (Ala) in B. afzelii. B. garinii has more variation and in addition to the previous two differences, having at least one more difference, where in B. burgdorferi is a Lysine (Lys), and sometimes also has a deletion at B. burgdorferi’s Alanine 208. LA-2 and OspA of B. burgdorferi form a tight interface when binding, and the longer Glutamine (Gln) sidechain found in B. afzelii and B. garinii is more difficult to accommodate, causing less binding. A chimera that was weakly recognized by LA-2 was made with parts of loop 1 from B. burgdorferi, and loops 2 and 3 from B. garinii.[23] Recently, a different kind of chimera has been made which combined the proximal region of B. burgdorferi and distal region of B. afzelii, and was able to successfully protect mice from both species.[31]
Acute Lyme Neuroborreliosis (LNB)
Acute Lyme Neuroborreliosis (LNB) is part of a later stage of Lyme disease in which the spirochete invades the peripheral and central nervous systems (CNS). Symptoms of LNB include: meningoradiculitis with inflammation of the nerve roots and radiculitis (Bannwarth’s syndrome), lymphocytic meningitis, and cranial and peripheral neuritis. In Europe, the strain predominantly found in the CSF of patients with Bannwarth's syndrome is B. garinii. However, in the United States, Bannwarth's syndrome is rare and the most common manifestations of Lyme neuroborreliosis is meningitis, caused by B. burgdorferi. The presence of OspA in the cerebrospinal fluid (CSF) is responsible for this complex inflammatory response in the brain that leads to the neuroborreliosis.[29]
It is not fully understood how B. burgdorferi get past the blood-brain barrier, though some researchers suggest a paracellular route, which involves a process using transient tether-type associations involving OspA. The blood-brain barrier is composed of brain microvascular endothelial cells, among other cells. Studies have shown that OspA adheres to brain microvascular cells by binding to the CD40 receptors, which is followed by an induction of signaling cascades, and adhesion to endothelial cells, resulting in the movement of B. burgdorferi into the CNS. Similar cell signaling events are seen when leukocytes cross the blood brain barrier, and it has been proposed that B. burgdorferi may mimic this process. It has been found that not all strains of B. burgdorferi can utilize OspA to cross into the CNS, and OspA only contributes about 70% to adherence, and other B. burgdorferi proteins are also needed in this process. It has also been seen that OspA mediates the adhesion of B. burgdorferi to murine neural and glial cell lines. [32]
There are many steps involved in the host's inflammatory response to OspA. When B. burgdorferi enter the host’s CNS they encounter several different types of immune cells such as monocytes, macrophages, and dendritic cells. While in the CSF, OspA is upregulated and it’s increased expression promotes recognition by immune cells, such as monocytes. Upon recognition of OspA, monocytes release proinflammatory cytokines (i.e. interferon), as well as chemokines, such as CXCL13. In patients with LNB, there is an observed increase in the levels of these cytokines and chemokines in their CSF. The production of chemokines leads to the recruitment of other immune cells to the site of infection. B-lymphocytes respond to the new concentration gradient of CXCL13 and other chemokines between the blood and CSF and migrate into the CSF. The B-lymphocytes then differentiate and mature into plasma cells. The plasma cells create large quantities of anti-OspA antibodies specific to this strain of B. burgdorferi and release them into the CSF to target the pathogen[29]. This process is two-sided in the sense that the OspA aids in the onset of new symptoms (neuroborreliosis) through the chemokine’s actions, as well as initiating the signaling cascade to destroy itself.
Evasion and the Extracellular Matrix
The B. burgdorferi are able to hide in the extracellular matrix, allowing it to survive by avoiding leukocytes circulating in the bloodstream. OspA can rapidly bind to plasminogen, which facilitates the spread of the bacteria[33][34]. By binding to plasminogen, B. burgdorferi could be exploiting its function and utilizing it to invade the extracellular matrix. However, due to the fact that OspA is downregulated during feeding, and stays unexpressed, a different mechanism may be used instead. Additionally, B. burgdorferi induces the local upregulation of matrix metalloproteinase-9, causing the digestion of the surrounding extracellular matrix. B. burgdorferi can also bind to several proteins in the extracellular matrix, such as fibronectin, integrins or decorin, which can aid in the spread and survival of the spirochetes in these tissues.[29]
Antibodies to OspA
Interaction between OspA and LA-2
LA-2 is an IgM murine monoclonal antibody that is being used in vaccine development that interacts with on the C-terminal of OspA, as described above. These interactions include eight direct hydrogen bonds, four solvent-bridged hydrogen bonds, three ion pairs, and numerous van der Waals interactions.[23]
Structural changes to OspA in the complexed form
Conformational changes upon the binding of OspA and LA-2 show that LA-2 recognition of OspA involves an induced fit mechanism where the conformations of loops 1-3 shift to optimize complementarity to the antigen-combining site.[23] The overall structure of the C-terminal of OspA is unchanged upon the binding of LA-2 with comparison to the free OspA. The maximum atomic shift is 4.7Å at the site of .[23]
OspA-based Vaccine
The membrane composition of B. burgdorferi is abundant in both OspA and OspB, and the two proteins share a 53% similarity in their primary sequences, however, OspB has greater variability than OspA.[16]. Of the three exposed loops found on OspA, only loop 1 is variable while loops 2 and 3 are conserved. This makes OspA a more consistent antigen (compared to OspB and OspC) for the immune system to target and usable as a vaccine to Lyme disease.[23] The first vaccine developed against Lyme disease, Lymerix, used a purified recombinant form of OspA and functioned in blocking transmission of the spirochetes expressing OspA from tick to host during feeding, killing them while still attached to the tick's gut.[3][35] The vaccine was 76% to 92% effective in separate clinical trials in which patients were treated for two years following a three-dose schedule. However, the vaccine was suspended from use in 2002 when opponents claimed the IgG antibodies for OspA were associated with the onset of severe chronic arthritis, as well as other side effects affecting immunity.[3][36] This claim, in conjunction with the desire for a more widespread vaccine treating multiple strains of B. burgdorferi, has spurred research towards a new vaccine. One goal is to develop a vaccine with broader protection, creation of a chimera, mixing the OspA of different strains of B. burgdorferi.
Study of the epitope of OspA and its interactions with the murine monoclonal antibody LA-2 have proved useful in determining effectiveness of a given vaccine trial as high levels of antibodies in test sera compete against LA-2 for binding with OspA. LA-2 makes direct contact with three exposed loops of the C-terminus of OspA. The recognition of OspA by LA-2 requires an induced fit mechanism where these three loops undergo conformational changes to optimize their interaction in the complex. [23]
VlsE and Lyme Disease
| |
| Variable Major Protein (VMP)-like sequence Expressed (1l8w), resolution 2.3Å

- This representation of VlsE illustrates the only crystal structure available on the PDB site. There are 1030 possible combinations of the VR.
|
Variable Major Protein -like sequence Expressed (VlsE) is another surface lipoprotein of B. burgdorferi. It undergoes antigenic variation seemingly important in evasion of the host’s immune system. In addition, the protein is used for Lyme disease diagnosis.
Structure of VlsE
VlsE is composed of four similar subunits each possessing two invariable domains and one variable domain.[37] The variable domain contains six variable regions (VR1-VR6), and six invariable regions (IR1-IR6). Research suggests that the protein may exist as a dimer where each monomeric C & N termini neighbor each other forming the membrane proximal portion of the protein, and the variable regions form the membrane distal portion.[38] [39] The invariable regions are largely embedded in the protein and remain relatively unchanged within the host and across strains. The variable regions encompass 37% of the VlsE’s exposed surface area despite comprising only 25% of the protein.[37] [38] However, 50% of the VR surface area is exposed while IR6, a strong antigen, exposes just 13.7% of its surface area. This leaves only of the antigenic IR6 unprotected: lysine-276, glutamine-279, lysine-291, and lysine-294. Thus, it is almost entirely embedded in the protein and .[38]
Antigenicity
The variable regions undergo a recombination event stimulated by the host’s cytokines while absence of those cytokines results in a decreased bacterial burden.[40] Recombination leads to variation with an estimated 1030 possible combinations, far exceeding the number of antibodies found in the human immune system. While the VR does exhibit antigenicity, this recombination makes it unlikely that a sufficient amount of a single VR variation will be present in large enough supply to lead to an immunodominant variable region.[41] IR6, however, exhibits immunodominance while IR1-IR5 are primarily nonantigenic in humans. Thus, shielding of the immunodominant IR6 by VR regions not subject to antibody response allows for IR6 to elicit an immune response while remaining inaccessible to antibody binding. Research suggests that the 26 amino residues of may function as a single epitope with a central alpha helical core.[38] [40] [42]
Function in Immune System Evasion
VlsE is essential to the persistence and virulence of Lyme disease and is upregulated under humoral immune pressure.[43] [44] While the exact mechanism for immune evasion remains unknown, several theories have been put forth. One popular theory maintains that VlsE masks other surface antigens by coating the surface of the bacteria, thereby sterically blocking the antigens from antibody binding. This is similar to other pathogens with variable regions, such as Trypanosoma brucei, the protozoa responsible for African sleeping sickness and Neisseria gonorrhea, the bacterial cause of gonorrhea. However, recent studies have cast doubt on this theory. An alternate theory provides that VlsE directly stimulates B cell antibody production independent of T-cells. The robust response elicited is thought to override antibody production against other antigens.[43]
C6 Diagnostic Testing
Throughout the course of the disease, IR6 produces a strong antibody response that can be identified from early to late phases. Applications in diagnostic testing have been identified as a result of this strong immune response and IR6’s relative invariability across strains.[45] [42] A C6 ELISA test has been developed which uses a 26 amino acid synthetic peptide, C6, containing the IR6 sequence. Results show 99% specificity and 100% precision with high sensitivity. In fact, OspA vaccination does not influence C6 specificity; therefore, C6 ELISA tests are valuable diagnostic tools.[45] The CDC currently recommends a two-step test incorporating first a polyvalent, whole-cell sonicate (WCS) immunofluorescent assay. If results are positive, this is followed by IgG and IgM WCS Western blots to eliminate false positives.[46] Therefore, this one-step ELISA test presents an accurate and economical alternative to the current two-step model.[45]
References
- ↑ Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982 Jun 18;216(4552):1317-9. PMID:7043737
- ↑ Benach JL, Bosler EM, Hanrahan JP, Coleman JL, Habicht GS, Bast TF, Cameron DJ, Ziegler JL, Barbour AG, Burgdorfer W, Edelman R, Kaslow RA. Spirochetes isolated from the blood of two patients with Lyme disease. N Engl J Med. 1983 Mar 31;308(13):740-2. PMID:6828119 doi:http://dx.doi.org/10.1056/NEJM198303313081302
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Connolly SE, Benach JL. The versatile roles of antibodies in Borrelia infections. Nat Rev Microbiol. 2005 May;3(5):411-20. PMID:15864264 doi:10.1038/nrmicro1149
- ↑ Templeton TJ. Borrelia outer membrane surface proteins and transmission through the tick. J Exp Med. 2004 Mar 1;199(5):603-6. Epub 2004 Feb 23. PMID:14981110 doi:10.1084/jem.20040033
- ↑ Templeton TJ. Borrelia outer membrane surface proteins and transmission through the tick. J Exp Med. 2004 Mar 1;199(5):603-6. Epub 2004 Feb 23. PMID:14981110 doi:10.1084/jem.20040033
- ↑ 6.0 6.1 6.2 Kumaran D, Eswaramoorthy S, Luft BJ, Koide S, Dunn JJ, Lawson CL, Swaminathan S. Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi. EMBO J. 2001 Mar 1;20(5):971-8. PMID:11230121 doi:http://dx.doi.org/10.1093/emboj/20.5.971
- ↑ 7.0 7.1 7.2 7.3 Brisson D, Dykhuizen DE. A modest model explains the distribution and abundance of Borrelia burgdorferi strains. Am J Trop Med Hyg. 2006 Apr;74(4):615-22. PMID:16606995
- ↑ 8.0 8.1 Earnhart CG, Marconi RT. An octavalent lyme disease vaccine induces antibodies that recognize all incorporated OspC type-specific sequences. Hum Vaccin. 2007 Nov-Dec;3(6):281-9. Epub 2007 Jul 2. PMID:17921702
- ↑ 9.0 9.1 Eicken C, Sharma V, Klabunde T, Owens RT, Pikas DS, Hook M, Sacchettini JC. Crystal structure of Lyme disease antigen outer surface protein C from Borrelia burgdorferi. J Biol Chem. 2001 Mar 30;276(13):10010-5. Epub 2001 Jan 3. PMID:11139584 doi:10.1074/jbc.M010062200
- ↑ Earnhart C, LeBlanc D, Alix K, Desrosiers D, Radolf J, and Marconi R. 2010. Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Molecular Microbiology 76(2): 393-408. DOI: 10.1111/j.1365-2958.2010.07103.x
- ↑ LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci U S A. 2003 Jan 21;100(2):567-71. Epub 2003 Jan 13. PMID:12525705 doi:10.1073/pnas.0233733100
- ↑ Brisson D, Dykhuizen DE. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics. 2004 Oct;168(2):713-22. PMID:15514047 doi:10.1534/genetics.104.028738
- ↑ Christopher G. Earnhart, Eric L. Buckles, Richard T. Marconi. "Development of an OspC-based tetravalent, recombinant, chimeric vaccinogen that elicits bactericidal antibody against diverse Lyme disease spirochete strains, Vaccine." 25(3) 466-480 (2007). DOI: 10.1016/j.vaccine.2006.07.052
- ↑ LaRocca TJ, Benach JL. The important and diverse roles of antibodies in the host response to Borrelia infections. Curr Top Microbiol Immunol. 2008;319:63-103. PMID:18080415
- ↑ Neelakanta G, Li X, Pal U, Liu X, Beck DS, DePonte K, Fish D, Kantor FS, Fikrig E. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog. 2007 Mar;3(3):e33. PMID:17352535 doi:10.1371/journal.ppat.0030033
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 16.7 Becker M, Bunikis J, Lade BD, Dunn JJ, Barbour AG, Lawson CL. Structural investigation of Borrelia burgdorferi OspB, a bactericidal Fab target. J Biol Chem. 2005 Apr 29;280(17):17363-70. Epub 2005 Feb 15. PMID:15713683 doi:10.1074/jbc.M412842200
- ↑ Meri T, Cutler SJ, Blom AM, Meri S, Jokiranta TS. Relapsing fever spirochetes Borrelia recurrentis and B. duttonii acquire complement regulators C4b-binding protein and factor H. Infect Immun. 2006 Jul;74(7):4157-63. PMID:16790790 doi:10.1128/IAI.00007-06
- ↑ Pietikainen J, Meri T, Blom AM, Meri S. Binding of the complement inhibitor C4b-binding protein to Lyme disease Borreliae. Mol Immunol. 2010 Mar;47(6):1299-305. doi: 10.1016/j.molimm.2009.11.028. Epub, 2009 Dec 21. PMID:20022381 doi:10.1016/j.molimm.2009.11.028
- ↑ LaRocca TJ, Benach JL. The important and diverse roles of antibodies in the host response to Borrelia infections. Curr Top Microbiol Immunol. 2008;319:63-103. PMID:18080415
- ↑ . PMID:216315890657
- ↑ Coleman JL, Rogers RC, Benach JL. Selection of an escape variant of Borrelia burgdorferi by use of bactericidal monoclonal antibodies to OspB. Infect Immun. 1992 Aug;60(8):3098-104. PMID:1639477
- ↑ Coleman JL, Rogers RC, Rosa PA, Benach JL. Variations in the ospB gene of Borrelia burgdorferi result in differences in monoclonal antibody reactivity and in production of escape variants. Infect Immun. 1994 Jan;62(1):303-7. PMID:7505260
- ↑ 23.0 23.1 23.2 23.3 23.4 23.5 23.6 23.7 23.8 Ding W, Huang X, Yang X, Dunn JJ, Luft BJ, Koide S, Lawson CL. Structural identification of a key protective B-cell epitope in Lyme disease antigen OspA. J Mol Biol. 2000 Oct 6;302(5):1153-64. PMID:11183781 doi:10.1006/jmbi.2000.4119
- ↑ 24.0 24.1 Escudero R, Halluska ML, Backenson PB, Coleman JL, Benach JL. Characterization of the physiological requirements for the bactericidal effects of a monoclonal antibody to OspB of Borrelia burgdorferi by confocal microscopy. Infect Immun. 1997 May;65(5):1908-15. PMID:9125579
- ↑ Schwan TG, Schrumpf ME, Karstens RH, Clover JR, Wong J, Daugherty M, Struthers M, Rosa PA. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J Clin Microbiol. 1993 Dec;31(12):3096-108. PMID:8308101
- ↑ Benjamin DC, Williams DC Jr, Smith-Gill SJ, Rule GS. Long-range changes in a protein antigen due to antigen-antibody interaction. Biochemistry. 1992 Oct 13;31(40):9539-45. PMID:1382591
- ↑ Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002 Dec;102(12):4501-24. PMID:12475199
- ↑ Pal U, Li X, Wang T, Montgomery RR, Ramamoorthi N, Desilva AM, Bao F, Yang X, Pypaert M, Pradhan D, Kantor FS, Telford S, Anderson JF, Fikrig E. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell. 2004 Nov 12;119(4):457-68. PMID:15537536 doi:10.1016/j.cell.2004.10.027
- ↑ 29.0 29.1 29.2 29.3 Rupprecht TA, Koedel U, Fingerle V, Pfister HW. The pathogenesis of lyme neuroborreliosis: from infection to inflammation. Mol Med. 2008 Mar-Apr;14(3-4):205-12. PMID:18097481 doi:10.2119/2007-00091.Rupprecht
- ↑ Makabe K, Tereshko V, Gawlak G, Yan S, Koide S. Atomic-resolution crystal structure of Borrelia burgdorferi outer surface protein A via surface engineering. Protein Sci. 2006 Aug;15(8):1907-14. Epub 2006 Jul 5. PMID:16823038 doi:10.1110/ps.062246706
- ↑ Livey I, O'Rourke M, Traweger A, Savidis-Dacho H, Crowe BA, Barrett PN, Yang X, Dunn JJ, Luft BJ. A new approach to a Lyme disease vaccine. Clin Infect Dis. 2011 Feb;52 Suppl 3:s266-70. PMID:21217174 doi:10.1093/cid/ciq118
- ↑ Pulzova L, Kovac A, Mucha R, Mlynarcik P, Bencurova E, Madar M, Novak M, Bhide M. OspA-CD40 dyad: ligand-receptor interaction in the translocation of neuroinvasive Borrelia across the blood-brain barrier. Sci Rep. 2011;1:86. Epub 2011 Sep 8. PMID:22355605 doi:10.1038/srep00086
- ↑ Coleman JL, Sellati TJ, Testa JE, Kew RR, Furie MB, Benach JL. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect Immun. 1995 Jul;63(7):2478-84. PMID:7790059
- ↑ Coleman JL, Gebbia JA, Piesman J, Degen JL, Bugge TH, Benach JL. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell. 1997 Jun 27;89(7):1111-9. PMID:9215633
- ↑ Battisti JM, Bono JL, Rosa PA, Schrumpf ME, Schwan TG, Policastro PF. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect Immun. 2008 Nov;76(11):5228-37. Epub 2008 Sep 8. PMID:18779341 doi:10.1128/IAI.00410-08
- ↑ Plotkin SA. Correcting a public health fiasco: The need for a new vaccine against Lyme disease. Clin Infect Dis. 2011 Feb;52 Suppl 3:s271-5. PMID:21217175 doi:10.1093/cid/ciq119
- ↑ 37.0 37.1 Liang FT, Philipp MT. Analysis of antibody response to invariable regions of VlsE, the variable surface antigen of Borrelia burgdorferi. Infect Immun. 1999 Dec;67(12):6702-6. PMID:10569796
- ↑ 38.0 38.1 38.2 38.3 Eicken C, Sharma V, Klabunde T, Lawrenz MB, Hardham JM, Norris SJ, Sacchettini JC. Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia burgdorferi. J Biol Chem. 2002 Jun 14;277(24):21691-6. Epub 2002 Mar 28. PMID:11923306 doi:10.1074/jbc.M201547200
- ↑ Jones K, Guidry J, Wittung-Stafshede P. Characterization of surface antigen from Lyme disease spirochete Borrelia burgdorferi. Biochem Biophys Res Commun. 2001 Nov 30;289(2):389-94. PMID:11716485 doi:10.1006/bbrc.2001.5983
- ↑ 40.0 40.1 Anguita J, Thomas V, Samanta S, Persinski R, Hernanz C, Barthold SW, Fikrig E. Borrelia burgdorferi-induced inflammation facilitates spirochete adaptation and variable major protein-like sequence locus recombination. J Immunol. 2001 Sep 15;167(6):3383-90. PMID:11544329
- ↑ Liang FT, Alvarez AL, Gu Y, Nowling JM, Ramamoorthy R, Philipp MT. An immunodominant conserved region within the variable domain of VlsE, the variable surface antigen of Borrelia burgdorferi. J Immunol. 1999 Nov 15;163(10):5566-73. PMID:10553085
- ↑ 42.0 42.1 Liang FT, Philipp MT. Epitope mapping of the immunodominant invariable region of Borrelia burgdorferi VlsE in three host species. Infect Immun. 2000 Apr;68(4):2349-52. PMID:10722641
- ↑ 43.0 43.1 Bankhead T, Chaconas G. The role of VlsE antigenic variation in the Lyme disease spirochete: persistence through a mechanism that differs from other pathogens. Mol Microbiol. 2007 Sep;65(6):1547-58. Epub 2007 Aug 21. PMID:17714442 doi:10.1111/j.1365-2958.2007.05895.x
- ↑ Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, Fikrig E. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun. 2004 Oct;72(10):5759-67. PMID:15385475 doi:10.1128/IAI.72.10.5759-5767.2004
- ↑ 45.0 45.1 45.2 Liang FT, Steere AC, Marques AR, Johnson BJ, Miller JN, Philipp MT. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi vlsE. J Clin Microbiol. 1999 Dec;37(12):3990-6. PMID:10565920
- ↑ Branda JA, Linskey K, Kim YA, Steere AC, Ferraro MJ. Two-tiered antibody testing for Lyme disease with use of 2 enzyme immunoassays, a whole-cell sonicate enzyme immunoassay followed by a VlsE C6 peptide enzyme immunoassay. Clin Infect Dis. 2011 Sep;53(6):541-7. PMID:21865190 doi:10.1093/cid/cir464
Teaching at Stony Brook University
This Proteopedia page is the product of a new introductory biology laboratory started in the spring of 2012 at Stony Brook University. Undergraduates model and print tactile 3-dimensional proteins involved in Lyme disease in order to understand and interpret contemporary structural biology research. The best student-authored summaries from spring and summer 2012 were selected for this Proteopedia page, thereby connecting students to scientists and facilitating further research experiences.
Author contributions:
OspC: Irene Chen, Khine Tun
Antibodies to Osp A and B: Safa Abdelhakim, Alexandros Konstantinidis, Philip J. Pipitone, Christopher Smilios
OspA: Jenny Kim Kim, Cara Lin, Andrea Mullen, Kimberly Slade
OspB: Olivia Cheng, Stephanie Maung, Ying Zhao
VlsE: Frank J. Albergo, Rachel Cirineo, Tanya Turkewitz
Editors, teachers: Jeff Ecklund, Joan M. Miyazaki, Christopher Morales, Carol Nicosia, Deborah A. Spikes, Raymond Suhandynata, La Zhong, Jonathan Manit Wyrick
Technical support: Nancy A. Black, Jameson T. Crowley
Collaborating research scientists, editors: Jorge L. Benach, Timothy J. LaRocca
Course co-developer, writer, editor: Niamh B. O'Hara
Course director, course co-developer, writer, editor: Marvin H. O'Neal III
Supported by: Howard Hughes Medical Institute 52006940
Proteopedia Page Authors
Safa Abdelhakim,
Frank J. Albergo,
Irene Chen,
Olivia Cheng,
Rachel Cirineo,
Jenny Kim Kim,
Alexandros Konstantinidis,
Cara Lin,
Stephanie Maung,
Christopher Morales,
Andrea Mullen,
Niamh B. O'Hara,
Marvin H. O'Neal III,
Philip J. Pipitone,
Kimberly Slade,
Christopher Smilios,
Raymond Suhandynata,
Khine Tun,
Tanya Turkewitz,
Ying Zhao,
La Zhong,
Jonathan Manit Wyrick.
|



![Figure 3: A Diagram of the Life Cycle of the Blacklegged Tick.[[1]]](/wiki/images/thumb/7/7d/Life_cycle_of_tick.png/300px-Life_cycle_of_tick.png)
![Figure 4: Illustrated Prevalence of Lyme Disease in the United States (Generated by the CDC).[[2]]](/wiki/images/thumb/9/9d/Lyme_Disease_Risk_Map.gif/300px-Lyme_Disease_Risk_Map.gif)



