Sandbox Reserved 921
From Proteopedia
| Line 25: | Line 25: | ||
==Structure== | ==Structure== | ||
| - | The <scene name='57/573136/Starting_view/1'>surface</scene> of FAAH reveals two openings directly accessible by the inner layer of the [http://en.wikipedia.org/wiki/Lipid_bilayer lipid bilayer].<ref name= "1MT5">PMID:12459591</ref> These <scene name='57/573136/Membrane_access_channel/ | + | The <scene name='57/573136/Starting_view/1'>surface</scene> of FAAH reveals two equivalent openings (<scene name='57/573136/Starting_view/4'>Opening 1</scene>, <scene name='57/573136/Starting_view/5'>Opening 2</scene>) directly accessible by the inner layer of the [http://en.wikipedia.org/wiki/Lipid_bilayer lipid bilayer].<ref name= "1MT5">PMID:12459591</ref> These <scene name='57/573136/Membrane_access_channel/8'>Membrane Access Channels</scene> (MAC) are made up of three flaps and two intrusions which collectively form the entry way for the aliphatic binding of the amide lipid substrate. Flaps 1 and 2 envelope the middle and backside of the anandamide mimic, and are locked together by a <scene name='57/573136/Membrane_access_channel/9'>salt bridge</scene> between Arg486 and Asp403. Flap 2 contains a very <scene name='57/573136/Membrane_access_channel/12'>hydrophobic membrane binding cap</scene> that partially covers the opening with Phe432. This binding cap clings to the cell's hydrophobic inner membrane and uses a multitude of <scene name='57/573136/Membrane_access_channel/13'>positively charged residues</scene> to lure out partitioned anandamide by its narrow partial negative charge. The catalytic site is defined by the catalytic triad: the <scene name='57/573136/Membrane_access_channel/14'>238-241 anionic hole loop</scene> contributes the nucleophilic S241, loop 3 contributes the neighboring <scene name='57/573136/Membrane_access_channel/15'>S217</scene> upon forming the very top of the membrane access channel, and a fourth loop contributes the K142. |
| + | |||
| + | The membrane access channel leads to the active site, which is flanked by both the <scene name='57/573136/Cytosolic_port/2'>acyl chain binding pocket and cytosolic port </scene> (ABP and CP).<ref name= "3LJ7">PMID:20493882</ref> The cytosolic port is a lengthy, flexible loop that leads directly into the [http://en.wikipedia.org/wiki/Cytoplasm cytoplasm], allowing the deacylated amine to enter the cell. | ||
Revision as of 16:24, 25 April 2014
| This Sandbox is Reserved from Jan 06, 2014, through Aug 22, 2014 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 911 through Sandbox Reserved 922. |
To get started:
More help: Help:Editing |
Fatty Acid Amide Hydrolase
Introduction
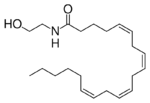
Fatty acid amide hydrolase (FAAH) is the primary catabolic enzyme for the degradation of fatty acid amides. FAAH primarily degrades anandamide, which is an endocannabinoid that activates the CB1 and CB2 cannabinoid receptors. When CB1 and CB2 cannabinoid receptors are active, the receptors affect appetite, sleep, and relief of pain. The ability to inhibit FAAH has been widely investigated for possible pain relief medication.
An inhibitor binds to the active site of the enzyme lowering the rate at which the enzyme degrades substrate, thus enzyme inhibitors are often used as drugs. A FAAH inhibitor offers great potential as a pain relief drug because FAAH commonly degrades anandamide. If the enzymatic activity of FAAH was lowered there would be increased levels of anandamide causing increased appetite, sleep, and pain relief.
A recent study on FAAH inhibitors combined an irreversible bond at Cys269 and a reversible bond at Ser241 of the active site.[1] A humanized rat variant of FAAH was inhibited by BR1, CL, and PEG. The mice displayed an increase in endogenous brain levels of the FAAH substrate anandamide for over six hours.[1] This is the first step towards developing a long lasting pain relief medication by inhibiting FAAH.
| |||||||||||
References
- ↑ 1.0 1.1 Otrubova K, Brown M, McCormick MS, Han GW, O'Neal ST, Cravatt BF, Stevens RC, Lichtman AH, Boger DL. Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues. J Am Chem Soc. 2013 Apr 24;135(16):6289-99. doi: 10.1021/ja4014997. Epub 2013 Apr, 12. PMID:23581831 doi:http://dx.doi.org/10.1021/ja4014997
- ↑ http://en.wikipedia.org/wiki/FAAH
- ↑ Kono M, Matsumoto T, Kawamura T, Nishimura A, Kiyota Y, Oki H, Miyazaki J, Igaki S, Behnke CA, Shimojo M, Kori M. Synthesis, SAR study, and biological evaluation of a series of piperazine ureas as fatty acid amide hydrolase (FAAH) inhibitors. Bioorg Med Chem. 2013 Jan 1;21(1):28-41. doi: 10.1016/j.bmc.2012.11.006. Epub, 2012 Nov 15. PMID:23218778 doi:http://dx.doi.org/10.1016/j.bmc.2012.11.006
- ↑ Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002 Nov 29;298(5599):1793-6. PMID:12459591 doi:10.1126/science.1076535
- ↑ Mileni M, Kamtekar S, Wood DC, Benson TE, Cravatt BF, Stevens RC. Crystal structure of fatty acid amide hydrolase bound to the carbamate inhibitor URB597: discovery of a deacylating water molecule and insight into enzyme inactivation. J Mol Biol. 2010 Jul 23;400(4):743-54. Epub 2010 May 21. PMID:20493882 doi:10.1016/j.jmb.2010.05.034