This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Lac repressor
From Proteopedia
| Line 1: | Line 1: | ||
<StructureSection load='1osl_19_1l1m_9_morph.pdb' size='450' side='right' scene='Morphs/1osl_19_1l1m_9_morph/2' caption=''> | <StructureSection load='1osl_19_1l1m_9_morph.pdb' size='450' side='right' scene='Morphs/1osl_19_1l1m_9_morph/2' caption=''> | ||
| - | + | __NOTOC__ | |
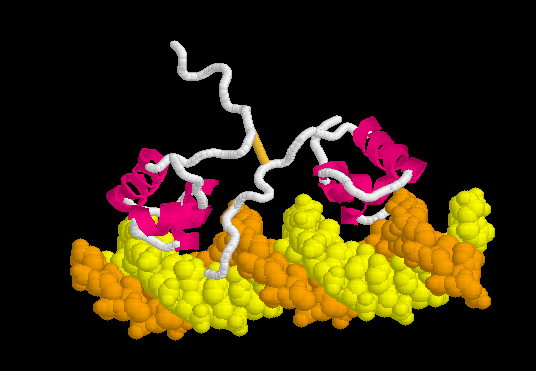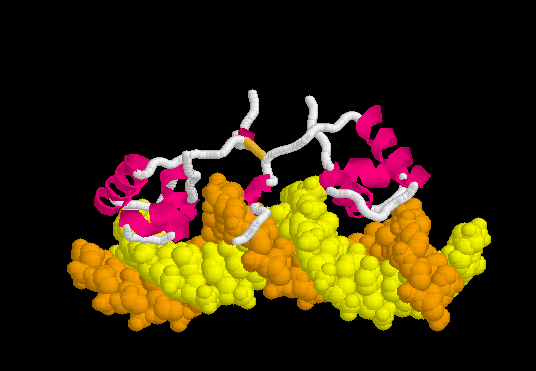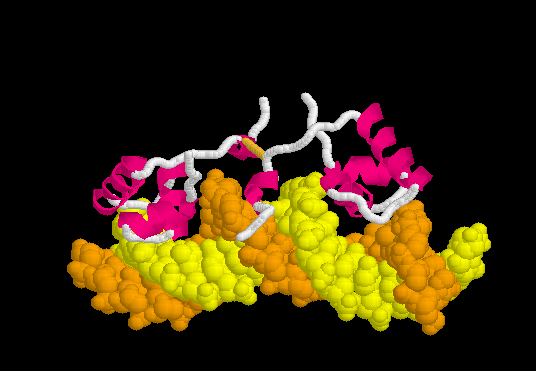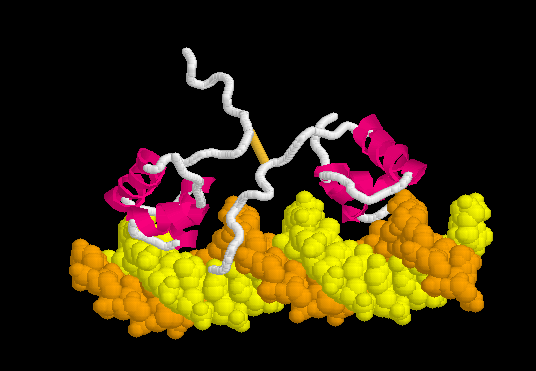
[[Morphs|Morph]] of the lac repressor complexed with DNA showing the differences between non-specific binding (straight DNA) vs. specific recognition of the operator sequence (kinked DNA). Whether the binding kinks the DNA, or simply stabilizes a pre-existing kink, is unknown. [[#Specific Binding| Details Below]]. | [[Morphs|Morph]] of the lac repressor complexed with DNA showing the differences between non-specific binding (straight DNA) vs. specific recognition of the operator sequence (kinked DNA). Whether the binding kinks the DNA, or simply stabilizes a pre-existing kink, is unknown. [[#Specific Binding| Details Below]]. | ||
| Line 44: | Line 44: | ||
DNA sequence recognition in the '''minor groove''', often accompanied by kinking or bending of the DNA, is more complex. Direct readout is less important, since, unlike in the major groove, the four bases do not present unique hydrogen-bonding surfaces in the minor groove<ref name="rohsrev2010" />. Recognition of the shape of the DNA seems more important<ref>PMID: 17981120</ref><ref name="rohs2009" />. In many cases, cationic arginines are believed to be attracted to a region of the minor groove with high aninoic charge density resulting from narrowing of the groove<ref name="rohs2009" >PMID: 19865164</ref>. In these cases, the protein appears to recognize the shape of the DNA minor groove (''indirect readout'')<ref name="rohs2009" />. | DNA sequence recognition in the '''minor groove''', often accompanied by kinking or bending of the DNA, is more complex. Direct readout is less important, since, unlike in the major groove, the four bases do not present unique hydrogen-bonding surfaces in the minor groove<ref name="rohsrev2010" />. Recognition of the shape of the DNA seems more important<ref>PMID: 17981120</ref><ref name="rohs2009" />. In many cases, cationic arginines are believed to be attracted to a region of the minor groove with high aninoic charge density resulting from narrowing of the groove<ref name="rohs2009" >PMID: 19865164</ref>. In these cases, the protein appears to recognize the shape of the DNA minor groove (''indirect readout'')<ref name="rohs2009" />. | ||
| - | + | In the lac repressor complex with specific DNA, a pair of arginines (Arg51 in each chain) is close to the minor groove, but points away from the groove (<scene name='Lac_repressor/Arg51/1'>restore initial scene</scene>). <!--[Remove this in view of <ref>PMID: 20232938</ref>? Also this portion of the minor groove does not contain ApT or ApA (TpT) which are associated with minor groove narrowing and high negative charge density.]--> Hence the binding of arginines to narrow minor grooves does not appear to be involved in specific DNA recognition by the lac repressor. | |
| - | In the lac repressor complex with specific DNA, a pair of arginines ( | + | |
====DNA Kinks==== | ====DNA Kinks==== | ||
| Line 85: | Line 84: | ||
Answers are available on request to {{Template:Contact}}. If you would like us to make the answers publically available within Proteopedia, please let us know. When contacting us, please give your full name, your position, institution or school, and location. | Answers are available on request to {{Template:Contact}}. If you would like us to make the answers publically available within Proteopedia, please let us know. When contacting us, please give your full name, your position, institution or school, and location. | ||
</StructureSection> | </StructureSection> | ||
| + | __NOTOC__ | ||
==Content Attribution & Acknowledgement== | ==Content Attribution & Acknowledgement== | ||
Revision as of 09:17, 4 May 2014
| |||||||||||
Content Attribution & Acknowledgement
The morphs displayed here were originally prepared by Eric Martz in 2004 for the page Lac Repressor Binding to DNA, within ProteinExplorer.Org.
Eric Martz thanks Remo Rohs for his kind and expert advice concerning the 2010-2011 updates to this article.
See Also
- Category: Lac repressor and Category: Lac Repressor, automatically-generated pages that list PDB codes for lac repressor models.
- Morphs where the morph of the lac repressor is used as an example.
- Lac repressor morph methods
- See: Regulation of Gene Expression for additional mechanisms of Gene Regulation
- For additional information, see: Transcription and RNA Processing
3D structures of Lac repressor
Updated on 04-May-2014
3edc – EcLAC + hexanediol - Escherichia coli
2pe5 – EcLAC residues 2-331 (mutant) + effector
1lbh - EcLAC + effector
2p9h - EcLAC residues 62-330 + effector
2paf - EcLAC residues 62-330 + anti-inducer
1lbi – EcLAC
1jye, 1jyf - EcLAC (mutant)
1lqc - EcLAC headpiece – NMR
1tlf - EcLAC residues 19-319
2r2v – LAC coiled-coil - yeast
Lac repressor complex with DNA
2kei, 1l1m – EcLAC DNA-binding domain (mutant) + O1 operator –NMR
2kej - EcLAC DNA-binding domain (mutant) + O2 operator – NMR
2kek - EcLAC DNA-binding domain (mutant) + O3 operator – NMR
2bjc - EcLAC DNA-binding domain (mutant) + GAL operator – NMR
1osl - EcLAC DNA-binding domain (mutant) + DNA – NMR
1cjg, 1lcc, 1lcd - EcLAC headpiece + DNA – NMR
1jwl - EcLAC + O1 operator + effector
1lbg - EcLAC + DNA + inducer
1efa - EcLAC residues 1-333 (mutant) + DNA
References & Notes
- ↑ L'opéron: groupe de gènes à expression coordonée par un opérateur. [Operon: a group of genes with the expression coordinated by an operator.] C R Hebd Seances Acad Sci., 250:1727-9, 1960. PubMed 14406329
- ↑ The lac repressor. Lewis, M. C R Biol. 328:521-48, 2005. PubMed 15950160
- ↑ This domain coloring scheme is adapted from Fig. 6 in the review by Lewis (C. R. Biol. 328:521, 2005). Domains are 1-45, 46-62, (63-162,291-320), (163-290,321-332), 330-339, and 340-357.
- ↑ Conservation results for 1lbg are from the precalculated ConSurf Database, using 103 sequences from Swiss-Prot with an average pairwise distance of 2.4.
- ↑ Conservation results for 1lbi are from the ConSurf Server, using 100 sequences from Uniprot with an average pairwise distance of 1.3.
- ↑ 6.0 6.1 For these scenes, the 20-model PDB files for 1osl and 1l1m were reduced in size, to avoid exceeding the java memory available to the Jmol applet. All atoms except amino acid alpha carbons and DNA phosphorus atoms were removed using the free program alphac.exe from PDBTools. Secondary structure HELIX records from the original PDB file header were retained. The results are Image:1osl ca.pdb and Image:1l1m ca.pdb.
- ↑ Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012 Jun 22;336(6088):1595-8. PMID:22723426 doi:10.1126/science.1221648
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233-69. PMID:20334529 doi:10.1146/annurev-biochem-060408-091030
- ↑ Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007 Nov 2;131(3):530-43. PMID:17981120 doi:10.1016/j.cell.2007.09.024
- ↑ 10.0 10.1 10.2 Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009 Oct 29;461(7268):1248-53. PMID:19865164 doi:10.1038/nature08473
- ↑ Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011 Feb 24;470(7335):498-502. Epub 2011 Jan 26. PMID:21270796 doi:10.1038/nature09775
- ↑ Honig B, Rohs R. Biophysics: Flipping Watson and Crick. Nature. 2011 Feb 24;470(7335):472-3. PMID:21350476 doi:10.1038/470472a
- ↑ Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010 Apr;17(4):423-9. Epub 2010 Apr 4. PMID:20364130 doi:10.1038/nsmb.1800
- ↑ Powerpoint is a registered trademark for a software package licensed by Microsoft Corp..
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Michal Harel, Alexander Berchansky, Joel L. Sussman, Karsten Theis, Henry Jakubowski, David Canner, Eran Hodis