Sandbox Reserved 930
From Proteopedia
| Line 23: | Line 23: | ||
==Introduction of the Myosin head S1 == | ==Introduction of the Myosin head S1 == | ||
<StructureSection load='1B7T' size='450' frame='true' side='right' caption='Myosin subfragment 1' scene='57/579700/Whole_structure/3'> | <StructureSection load='1B7T' size='450' frame='true' side='right' caption='Myosin subfragment 1' scene='57/579700/Whole_structure/3'> | ||
| - | Myosin is a large asymmetric molecule with a MW of about 500,000 kDa. It consist of | + | Myosin is a large asymmetric molecule with a MW of about 500,000 kDa. It consist of two globular head domains termed myosin subfragment 1 (S1), one neck subfragment 2 (S2) and a light meromyosin tail (LMM) <ref>PMID: 8203020</ref>. Myosin S1 unit comprises of a motor domain (MD) and a lever arm (Fig.3). By 2000 the structures of three scallop myosin S1 isoforms have been determined <ref>PMID: 11016966</ref><ref>PMID: 10338210</ref>, which are: |
• S1 nucleotide-free state corresponding to the rigor state of myosin, actin complex (PDB link 1DFK) | • S1 nucleotide-free state corresponding to the rigor state of myosin, actin complex (PDB link 1DFK) | ||
| Line 31: | Line 31: | ||
• S1 Mg-ADP state corresponding to the myosin detached state (1b7t) | • S1 Mg-ADP state corresponding to the myosin detached state (1b7t) | ||
| - | By comparing the available crystal structures of different myosin S1 unit isoforms, it enables us to understand the conformational changes within the motor domain depending on the nucleotide content in the active site. | + | |
| - | Here mainly the structure and function of MD relevant in the S1 Mg-ADP (pre power stroke) state will be discussed. | + | By comparing the available crystal structures of different myosin S1 unit isoforms, it enables us to understand the conformational changes within the motor domain depending on the nucleotide content in the active site. Here mainly the structure and function of MD relevant in the S1 Mg-ADP (pre power stroke) state will be discussed. |
Revision as of 11:34, 16 May 2014
| This Sandbox is Reserved from 01/04/2014, through 30/06/2014 for use in the course "510042. Protein structure, function and folding" taught by Prof Adrian Goldman, Tommi Kajander, Taru Meri, Konstantin Kogan and Juho Kellosalo at the University of Helsinki. This reservation includes Sandbox Reserved 923 through Sandbox Reserved 947. |
To get started:
More help: Help:Editing |
Contents |
Scallop myosin head in its pre power stroke state
Introduction
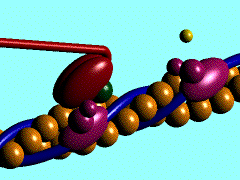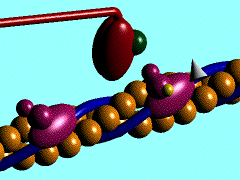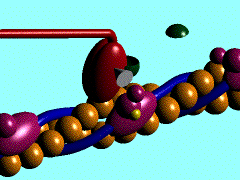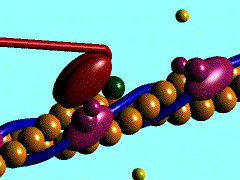
In the striated muscle the actin and myosin proteins form ordered basic units called sarcomeres. Muscle contraction is achieved by the mechanical sliding of myosin filament (thick filament) along the actin filament (thin filament), Fig. 1. The major constituent of the myosin filament is myosin, a motor protein responsible for converting chemical energy to mechanical movement. In the presence of Ca2+ and Mg2+, myosin is able to cyclically bind ATP and hydrolyse it to ADP + Pi , triggering subsequent myosin-actin detachment, reattachment and power stroke, so called contractile reaction (Fig.2).
.
Introduction of the Myosin head S1
| |||||||||||
References
1) Gourinath, S. et. al. 2003. Crystal Structure of Scallop Myosin S1 in the Pre-Power Stroke State to 2.6 Å Resolution: Flexibility and Function in the Head. Structure. 11(12): 1621–1627
2) Himmel, D. M. et. al. 2002. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: Further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 99(20): 12645–12650. 3) Houdusse, A. et. al. 2000. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. Oct 10, 2000; 97(21): 11238–11243.
4) Krans, J. 2010. The Sliding Filament Theory of Muscle Contraction. Nature Education 3(9):66
5) Risal, D. et. al. 2004. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci U S A. 101(24): 8930–8935.

