Sandbox Reserved 930
From Proteopedia
| Line 62: | Line 62: | ||
==Role of the subdomains and joints in the mechanism of the contractile cycle== | ==Role of the subdomains and joints in the mechanism of the contractile cycle== | ||
| - | The MD has different conformational states in each step of the contractile cycle. The conformation of the MD in each state depends on which nucleotide is bound to the active site (if any). In each structural state the conformation of the MD changes relatively little, but these changes are enough to cause a substantial difference in the position of the lever arm. | + | The MD has different conformational states in each step of the contractile cycle. The conformation of the MD in each state depends on which nucleotide is bound to the active site (if any). In each structural state the conformation of the MD changes relatively little, but these changes are enough to cause a substantial difference in the position of the lever arm <ref>PMID: 11016966</ref>. |
| - | The 50-kDa upper and lower subdomains as well as the converter control the motor function of the myosin head by rotating around the N-terminal subdomain. The rotations depend on the conformational changes of the 3 joints; switch II, SH1 helix region, and relay. The joints work together in the transition between the different conformational states of MD to control the overall organization of the myosin head. They also allow communication between the nucleotide-bonding pocket, acting-binding interface and the lever arm. | + | The 50-kDa upper and lower subdomains as well as the converter control the motor function of the myosin head by rotating around the N-terminal subdomain. The rotations depend on the conformational changes of the 3 joints; switch II, SH1 helix region, and relay. <ref>PMID: 15184651</ref>The joints work together in the transition between the different conformational states of MD to control the overall organization of the myosin head. They also allow communication between the nucleotide-bonding pocket, acting-binding interface and the lever arm <ref>PMID: 11016966</ref>. |
| - | Switch II, a catalytic loop of the nucleotide-binding pocket, moves in and out of the nucleotide-binding pocket during enzymatic activity. It is responsible for the unwinding of SH1 helix, along with the conformational changes caused by nucleotide binding. Upon unwinding helix SH1 uncouples the converter/lever module from the MD. Movement of the converter is controlled by the relay joint. The converter/relay module attains different conformations changing the position of the lever arm and thus giving rise to the different states of the actomyosin cycle. | + | Switch II, a catalytic loop of the nucleotide-binding pocket, moves in and out of the nucleotide-binding pocket during enzymatic activity. It is responsible for the unwinding of SH1 helix, along with the conformational changes caused by nucleotide binding. Upon unwinding helix SH1 uncouples the converter/lever module from the MD. <ref>PMID: 12297624</ref> Movement of the converter is controlled by the relay joint. The converter/relay module attains different conformations changing the position of the lever arm and thus giving rise to the different states of the actomyosin cycle <ref>PMID: 11016966</ref>. |
| - | Taking part in the organization of the different conformations of the contractile cycle is also the so called switch I, which is a second catalytic loop of the nucleotide-binding pocket. Switch II forms a specific salt bridge and hydrogen bond interactions with switch I that stabilize the pre-power stroke state. | + | Taking part in the organization of the different conformations of the contractile cycle is also the so called switch I, which is a second catalytic loop of the nucleotide-binding pocket. <ref>PMID: 11016966</ref>Switch II forms a specific salt bridge and hydrogen bond interactions with switch I that stabilize the pre-power stroke state <ref>PMID: 12297624</ref>. |
| + | |||
| + | In the pre-power stroke conformation of the MD, switch II interacts with the nucleotide-binding pocket and forms the stabilizing hydrogen bond interactions and a salt bridge with switch I. <ref>PMID: 12297624</ref> , <ref>PMID: 15184651</ref>Rotation of the 50-kDa upper subdomain away from the N-terminal subdomain pulls switches I and II apart breaking the protein- nucleotide interactions between switch I and ADP, as well as changing the conformation of switch II. These changes result in closing of the actin-binding site and opening of the nucleotide-binding pocket, leading to MgADP release. At the same timeSH1 helix is unwound and the lever arm is able to change its position enabling sliding of myosin through the actin filament <ref>PMID: 12297624</ref>. | ||
| - | In the pre-power stroke conformation of the MD, switch II interacts with the nucleotide-binding pocket and forms the stabilizing hydrogen bond interactions and a salt bridge with switch I. fig? Rotation of the 50-kDa upper subdomain away from the N-terminal subdomain pulls switches I and II apart breaking the protein- nucleotide interactions between switch I and ADP, as well as changing the conformation of switch II. These changes result in closing of the actin-binding site and opening of the nucleotide-binding pocket, leading to MgADP release. At the same timeSH1 helix is unwound and the lever arm is able to change its position enabling sliding of myosin through the actin filament. | ||
Revision as of 11:43, 16 May 2014
| This Sandbox is Reserved from 01/04/2014, through 30/06/2014 for use in the course "510042. Protein structure, function and folding" taught by Prof Adrian Goldman, Tommi Kajander, Taru Meri, Konstantin Kogan and Juho Kellosalo at the University of Helsinki. This reservation includes Sandbox Reserved 923 through Sandbox Reserved 947. |
To get started:
More help: Help:Editing |
Contents |
Scallop myosin head in its pre power stroke state
Introduction
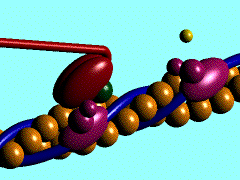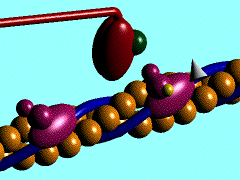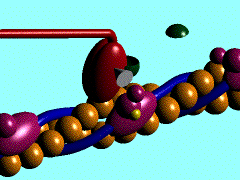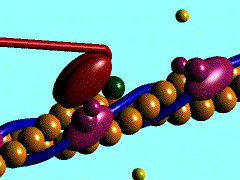
In the striated muscle the actin and myosin proteins form ordered basic units called sarcomeres. Muscle contraction is achieved by the mechanical sliding of myosin filament (thick filament) along the actin filament (thin filament), Fig. 1. The major constituent of the myosin filament is myosin, a motor protein responsible for converting chemical energy to mechanical movement. In the presence of Ca2+ and Mg2+, myosin is able to cyclically bind ATP and hydrolyse it to ADP + Pi , triggering subsequent myosin-actin detachment, reattachment and power stroke, so called contractile reaction (Fig.2).
.
Introduction of the Myosin head S1
| |||||||||||
References
1) Gourinath, S. et. al. 2003. Crystal Structure of Scallop Myosin S1 in the Pre-Power Stroke State to 2.6 Å Resolution: Flexibility and Function in the Head. Structure. 11(12): 1621–1627
2) Himmel, D. M. et. al. 2002. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: Further insights into the mechanics of the motor. Proc Natl Acad Sci U S A. 99(20): 12645–12650. 3) Houdusse, A. et. al. 2000. Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A. Oct 10, 2000; 97(21): 11238–11243.
4) Krans, J. 2010. The Sliding Filament Theory of Muscle Contraction. Nature Education 3(9):66
5) Risal, D. et. al. 2004. Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci U S A. 101(24): 8930–8935.

