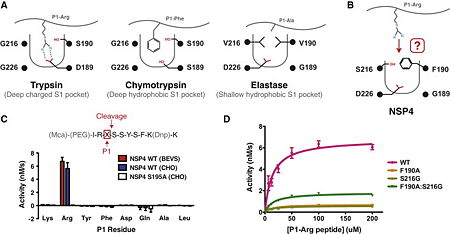
Isocitrate dehydrogenase
From Proteopedia
| Line 27: | Line 27: | ||
[[Isocitrate dehydrogenase]] (IDH) is an enzyme that is used during the third step of the [[Citric Acid Cycle|citric acid cycle]]. This biological reaction is an essential process that is used to create molecules that are used for cellular energy. In this step it catalyzes the oxidative decarboxylation of isocitrate meaning that CO2 is released from the isocitrate. In addition coenzyme [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NAD]+ is converted to an [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NADH]. This reaction results in an alpha-ketoglutarate molecule which is then moved on to the forth step of the citric acid cycle. In ''Escherichia coli'' the IDH is regulated by phosphorylation which is catalyzed by [[Isocitrate dehydrogenase kinase/phosphatase|IDH kinase/phosphatase (IDHK/P)]]. | [[Isocitrate dehydrogenase]] (IDH) is an enzyme that is used during the third step of the [[Citric Acid Cycle|citric acid cycle]]. This biological reaction is an essential process that is used to create molecules that are used for cellular energy. In this step it catalyzes the oxidative decarboxylation of isocitrate meaning that CO2 is released from the isocitrate. In addition coenzyme [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NAD]+ is converted to an [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NADH]. This reaction results in an alpha-ketoglutarate molecule which is then moved on to the forth step of the citric acid cycle. In ''Escherichia coli'' the IDH is regulated by phosphorylation which is catalyzed by [[Isocitrate dehydrogenase kinase/phosphatase|IDH kinase/phosphatase (IDHK/P)]]. | ||
| - | {{TOC limit|limit=2}} | ||
==Structural Analysis== | ==Structural Analysis== | ||
Isocitrate dehydrogenase is SCOP classified as an alpha beta structure. Its secondary composition consists of mainly alpha helices and beta sheets which are arranged into three layer alpha beta alpha <scene name='Michael_nobbe_sandbox_2/Alpha_beta_sandwich/1'>sandwich structures.</scene>. The entire protein consists of two side by side sandwich structures that face opposite directions. This then causes the proteins two active sites to face opposite directions. These two groups make up the A and B subunits of isocitrate dehydrogenase.In human isocitrate dehydrogenase there are 4 subunits. | Isocitrate dehydrogenase is SCOP classified as an alpha beta structure. Its secondary composition consists of mainly alpha helices and beta sheets which are arranged into three layer alpha beta alpha <scene name='Michael_nobbe_sandbox_2/Alpha_beta_sandwich/1'>sandwich structures.</scene>. The entire protein consists of two side by side sandwich structures that face opposite directions. This then causes the proteins two active sites to face opposite directions. These two groups make up the A and B subunits of isocitrate dehydrogenase.In human isocitrate dehydrogenase there are 4 subunits. | ||
| Line 33: | Line 32: | ||
==The Active Site== | ==The Active Site== | ||
The active site of Isocitrate dehydrogenase binds the NAD+ or an NADP+ molecule as well as an Mg2+ or Ca2+(<scene name='Michael_nobbe_sandbox_2/Isocitrate_active_site/4'>Active Site</scene>). This metal ion seems to be essential for catalysis<ref>PMID:15173171</ref>. Side chains of Asp 279 form hydrogen bonds with Ser94 (Human isocitrate dehydrogenase). Isocitrate is able to bind to the active site using about 8 amino acids like tyrosine, serine, asparagine, arginine, arginine, arginine, tyrosine, and lysine<ref>PMID:17632124</ref>. Across species these are not perfectly conserved but they are replaced with residues of similar properties and also are found in similar areas of the protein. Below is a picture of Porcine Active site with all of its residuals and ligands. | The active site of Isocitrate dehydrogenase binds the NAD+ or an NADP+ molecule as well as an Mg2+ or Ca2+(<scene name='Michael_nobbe_sandbox_2/Isocitrate_active_site/4'>Active Site</scene>). This metal ion seems to be essential for catalysis<ref>PMID:15173171</ref>. Side chains of Asp 279 form hydrogen bonds with Ser94 (Human isocitrate dehydrogenase). Isocitrate is able to bind to the active site using about 8 amino acids like tyrosine, serine, asparagine, arginine, arginine, arginine, tyrosine, and lysine<ref>PMID:17632124</ref>. Across species these are not perfectly conserved but they are replaced with residues of similar properties and also are found in similar areas of the protein. Below is a picture of Porcine Active site with all of its residuals and ligands. | ||
| - | |||
[[Image:Active site.jpg|left|450px|thumb]] | [[Image:Active site.jpg|left|450px|thumb]] | ||
| Line 49: | Line 47: | ||
[[Image:Mechanism.jpg]] | [[Image:Mechanism.jpg]] | ||
| - | |||
| - | |||
The mechanism by which isocitrate is converted to alpha-ketoglutarate <ref>http://en.wikipedia.org/wiki/File:IDHcatalyticmechanism.jpg</ref> | The mechanism by which isocitrate is converted to alpha-ketoglutarate <ref>http://en.wikipedia.org/wiki/File:IDHcatalyticmechanism.jpg</ref> | ||
| Line 71: | Line 67: | ||
</StructureSection> | </StructureSection> | ||
| - | __NOTOC__ | ||
==3D Structures of Isocitrate dehydrogenase== | ==3D Structures of Isocitrate dehydrogenase== | ||
Revision as of 06:58, 19 August 2014
| |||||||||||
Contents |
3D Structures of Isocitrate dehydrogenase
Updated on 19-August-2014
Isocitrate dehydrogenase
3blx – hIDH - yeast
2uxq – DpIDH – Desulfotalea psychrophila
3dms – IDH – Burkholderia pseudomallei
2e0c, 2dht - StIDH – Sulfolobus tokodaii
2iv0 – IDH – Archaeoglobus fulgidus
1zor – IDH – Thermotoga maritima
2b0t – IDH – Corynebacterium glutamicum
1xgv, 1v94 - ApIDH – Aeropyrum pernix
1pb3, 1sjs, 6icd, 7icd, 3icd – EcIDH – Escherichia coli
1idd, 1idf, 4icd - EcIDH (mutant)
3ah3 – TtIDH – Thermus thermophilus
3us8 – IDH – Sinorhizobium meliloti
4aoy – CtIDH – Clostridium thermocellum
IDH+citrate
2qfw – yIDH+isocitrate
3blv – yIDH+citrate
2uxr – DpIDH+isocitric acid
1itw – AvIDH+isocitrate+Mn – Azotobacter vinelandii
1lwd – IDH+isocitrate+Mn - pig
1pb1, 1p8f – EcIDH+isocitrate
1cw7, 5icd, 8icd - EcIDH +isocitrate+Mg
1cw1 - EcIDH (mutant)+isocitrate+Mn
1gro, 1grp - EcIDH (mutant)+isocitrate+Mg
1hqs – IDH+citrate – Bacillus subtilis
IDH+NADP
1t09 – hIDH+NADP - human
3mar – hIDH (mutant) + NADP
2qfv – yIDH+NADP
2e5m – StIDH+NADP
2cmj – mIDH+NADP – mouse
1tyo - ApIDH+NADP
1j1w – AvIDH+NADP – Azotobacter vinelandii
9icd - EcIDH +NADP
1iso - EcIDH (mutant)+NAD
3mbc - IDH (mutant)+NADP – Corynebacterium glutamicum
4hcx – IDH + NADP + Mn – mycobacterium tuberculosis
IDH+α-ketoglutarate
2qfy - yIDH+α-ketoglutarate
1cw4 – EcIDH (mutant)+α-ketoglutarate
1ika - EcIDH + α-ketoglutarate+Ca
3inm, 4l03, 4l04, 4l06 – hIDH (mutant)+NADPH+α-ketoglutarate+Ca – human
4kzo - hIDH +NADPH+α-ketoglutarate+Ca
2qfx - yIDH+NADPH+α-ketoglutarate+Ca
4ajc - EcIDH (mutant) +α-ketoglutarate+A2P+Ca
4ajr - EcIDH (mutant) +α-ketoglutarate+ NADP+ Mg
1bl5 - EcIDH +α-ketoglutarate+NADP+Mg
IDH binary complex
3asj – TtIDH + inhbibitor
IDH ternary complexes
3map - hIDH+NADP+isocitrate
3mas – hIDH (mutant) +NADP+isocitrate
1t0l – hIDH+NADP+isocitrate+Ca
4i3k, 4i3l, 4ja8 - hIDH (mutant) +NADP + inhibitor
3blw – yIDH+citrate+AMP
2cmv - mIDH+NADP+citrate
2d4v – IDH+NAD+citrate – Acidithiobacillus thiooxidans
2d1c – TtIDH+NADP+citrate
1xkd – ApIDH+NADP+citrate
1hj6 – EcIDH (mutant)+NADP+isopropylmalate+Mg
1idc - EcIDH (mutant)+oxalosuccinate+Mg
1ai3 - EcIDH +isocitrate+Mg+NDP
1ai2, 4aj3, 4aja - EcIDH +isocitrate+NADP+Ca
1ide, 4ajb - EcIDH (mutant)+isocitrate+Mg+NADP
4ajs - EcIDH (mutant)+isocitrate+Mg+ A2P + NMN
4aou - CtIDH + isocitrate+NADP+Mg
4aov - DpIDH + isocitrate+NADP+Mg
References
- ↑ Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004 Aug 6;279(32):33946-57. Epub 2004 Jun 1. PMID:15173171 doi:10.1074/jbc.M404298200
- ↑ Fedoy AE, Yang N, Martinez A, Leiros HK, Steen IH. Structural and functional properties of isocitrate dehydrogenase from the psychrophilic bacterium Desulfotalea psychrophila reveal a cold-active enzyme with an unusual high thermal stability. J Mol Biol. 2007 Sep 7;372(1):130-49. Epub 2007 Jun 19. PMID:17632124 doi:10.1016/j.jmb.2007.06.040
- ↑ http://en.wikipedia.org/wiki/Isocitrate_dehydrogenase#cite_note-nfr154197.2F32-6
- ↑ Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004 Aug 6;279(32):33946-57. Epub 2004 Jun 1. PMID:15173171 doi:10.1074/jbc.M404298200
- ↑ Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004 Aug 6;279(32):33946-57. Epub 2004 Jun 1. PMID:15173171 doi:10.1074/jbc.M404298200
- ↑ http://en.wikipedia.org/wiki/File:IDHcatalyticmechanism.jpg
- ↑ Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010 Dec 14;18(6):553-67. Epub 2010 Dec 9. PMID:21130701 doi:10.1016/j.ccr.2010.11.015
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Michael Nobbe, Joel L. Sussman