This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2emt
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | [[ | + | ==Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1== |
| + | <StructureSection load='2emt' size='340' side='right' caption='[[2emt]], [[Resolution|resolution]] 2.80Å' scene=''> | ||
| + | == Structural highlights == | ||
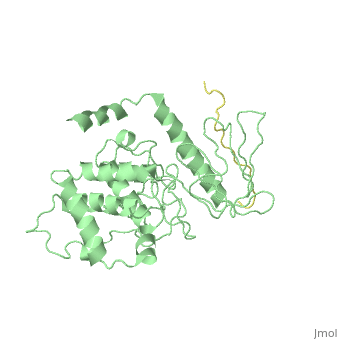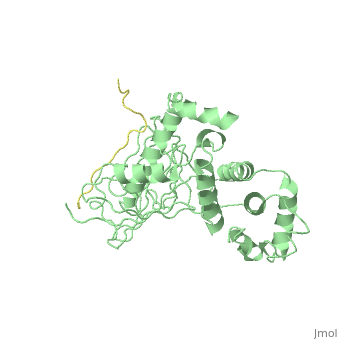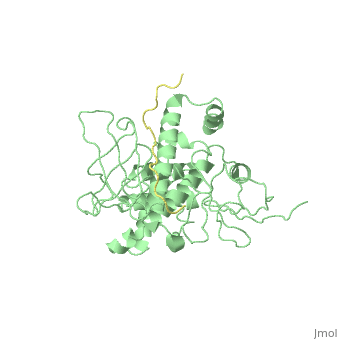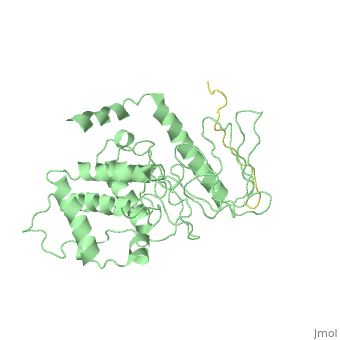
| + | <table><tr><td colspan='2'>[[2emt]] is a 5 chain structure with sequence from [http://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2EMT OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2EMT FirstGlance]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1j19|1j19]], [[1gc6|1gc6]], [[1gc7|1gc7]], [[2d11|2d11]], [[2d10|2d10]], [[2d2q|2d2q]], [[2ems|2ems]]</td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2emt FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2emt OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2emt RCSB], [http://www.ebi.ac.uk/pdbsum/2emt PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/em/2emt_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | P-selectin glycoprotein ligand-1 (PSGL-1), an adhesion molecule with O-glycosylated extracellular sialomucins, is involved in leukocyte inflammatory responses. On activation, ezrin-radixin-moesin (ERM) proteins mediate the redistribution of PSGL-1 on polarized cell surfaces to facilitate binding to target molecules. ERM proteins recognize a short binding motif, Motif-1, conserved in cytoplasmic tails of adhesion molecules, whereas PSGL-1 lacks Motif-1 residues important for binding to ERM proteins. The crystal structure of the complex between the radixin FERM domain and a PSGL-1 juxtamembrane peptide reveals that the peptide binds the groove of FERM subdomain C by forming a beta-strand associated with strand beta5C, followed by a loop flipped out towards the solvent. The Motif-1 3(10) helix present in the FERM-ICAM-2 complex is absent in PSGL-1 given the absence of a critical Motif-1 alanine residue, and PSGL-1 reduces its contact area with subdomain C. Non-conserved positions are occupied by large residues Met9 and His8, which stabilize peptide conformation and enhance groove binding. Non-conserved residues play an important role in compensating for loss of binding energy resulting from the absence of conserved residues important for binding. | ||
| - | + | Structural basis of PSGL-1 binding to ERM proteins.,Takai Y, Kitano K, Terawaki S, Maesaki R, Hakoshima T Genes Cells. 2007 Dec;12(12):1329-38. PMID:18076570<ref>PMID:18076570</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | + | ||
| - | == | + | |
| - | < | + | |
[[Category: Mus musculus]] | [[Category: Mus musculus]] | ||
[[Category: Hakoshima, T.]] | [[Category: Hakoshima, T.]] | ||
Revision as of 02:44, 30 September 2014
Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1
| |||||||||||