This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
VRC01 gp120 complex
From Proteopedia
| Line 11: | Line 11: | ||
HIV-1 enters its host by binding viral gp120, a surface glycoprotein of HIV, to the host cell’s CD4 receptor. This interaction induces conformational changes in gp120<ref name="wu" />. This conformational change results in the exposure of a binding site for the co-receptor, usually CCR5 OR CXCR4<ref>PMID: 21715490</ref>. The conformational changes also result in the formation of a pre-hairpin intermediate conformation in which gp41, a transmembrane glycoprotein of HIV, rearranges its molecules so that its N-terminal peptides form a trimer of helices that present a fusion peptide to the target cell. Once fusion occurs between the fusion peptide and the target cell membrane, HIV is able to enter and infect the target cell<ref>PMID: 22807678</ref>. VRC01 binds to CD4’s binding site on gp120, preventing the CD4 receptor from binding to HIV and infecting the cell<ref name="wu" />. | HIV-1 enters its host by binding viral gp120, a surface glycoprotein of HIV, to the host cell’s CD4 receptor. This interaction induces conformational changes in gp120<ref name="wu" />. This conformational change results in the exposure of a binding site for the co-receptor, usually CCR5 OR CXCR4<ref>PMID: 21715490</ref>. The conformational changes also result in the formation of a pre-hairpin intermediate conformation in which gp41, a transmembrane glycoprotein of HIV, rearranges its molecules so that its N-terminal peptides form a trimer of helices that present a fusion peptide to the target cell. Once fusion occurs between the fusion peptide and the target cell membrane, HIV is able to enter and infect the target cell<ref>PMID: 22807678</ref>. VRC01 binds to CD4’s binding site on gp120, preventing the CD4 receptor from binding to HIV and infecting the cell<ref name="wu" />. | ||
| - | + | ||
==Structural Features== | ==Structural Features== | ||
| + | <scene name='52/521911/Cv/3'>VRC01 heavy chain (gold, green, magenta, blue), light chain (aqua, pink, cyan, rust) in complex with gp120 (red, grey, yellow, wheat)</scene> ([[3ngb]]). | ||
| + | |||
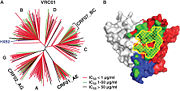
<u>Similarities to CD4 in complex with gp120</u>. Analysis of VRC01 in complex with gp120 shows that this complex covers 98% of the CD4 binding site. However, VRC01’s binding site extends outside that of CD4, making it vulnerable to resistance of VRC01 neutralization by antigenic variation<ref name="zhou">PMID: 20616231</ref>. [[Image: Target Site.jpg | thumb | alt=text | Picture 1]] In Picture 1B, the contact surface of VRC01 and CD4 are shown on gp120. The green represents VRC01's contact surface and yellow represents CD4's contact surface<ref name="kwong" />. Both the heavy chain and light chain of VRC01 contribute to the contact surfaces of the VRC01 gp120 complex. The focus of the binding is on the heavy chain second complementary-determining region. Over 50% of the surface contact involves the heavy chain second complementary-determining region; this is similar to CD4’s interaction with gp120. Two dominant residues, Phe43 and Arg59, are involved in CD4’s binding to gp120. Of these two residues, only the arginine interaction is mimicked by VRC01. This dominant interaction is between Asp368 of gp120 and Arg59 of the CD4 receptor and between Asp368 of gp120 and Arg71 of VRC01. Arg71 and Asp368 form a <scene name='VRC01_gp120_complex/Salt_bridge/1'>salt bridge</scene><ref name="zhou" />. | <u>Similarities to CD4 in complex with gp120</u>. Analysis of VRC01 in complex with gp120 shows that this complex covers 98% of the CD4 binding site. However, VRC01’s binding site extends outside that of CD4, making it vulnerable to resistance of VRC01 neutralization by antigenic variation<ref name="zhou">PMID: 20616231</ref>. [[Image: Target Site.jpg | thumb | alt=text | Picture 1]] In Picture 1B, the contact surface of VRC01 and CD4 are shown on gp120. The green represents VRC01's contact surface and yellow represents CD4's contact surface<ref name="kwong" />. Both the heavy chain and light chain of VRC01 contribute to the contact surfaces of the VRC01 gp120 complex. The focus of the binding is on the heavy chain second complementary-determining region. Over 50% of the surface contact involves the heavy chain second complementary-determining region; this is similar to CD4’s interaction with gp120. Two dominant residues, Phe43 and Arg59, are involved in CD4’s binding to gp120. Of these two residues, only the arginine interaction is mimicked by VRC01. This dominant interaction is between Asp368 of gp120 and Arg59 of the CD4 receptor and between Asp368 of gp120 and Arg71 of VRC01. Arg71 and Asp368 form a <scene name='VRC01_gp120_complex/Salt_bridge/1'>salt bridge</scene><ref name="zhou" />. | ||
<br/> | <br/> | ||
Revision as of 13:59, 18 December 2014
| |||||||||||
HIV Prevention Research
In 2003, Veazey and fellow researchers found that early broadly neutralizing antibodies had microbicide potential by using a monkey cell as the model. The microbicide used on these monkeys consisted of b12, a broadly neutralizing antibody. These monkeys were challenged with SHIV, simian-human immunodeficiency virus, through the vagina. Only three of the twelve monkeys became infected. It was also found that the protection against HIV lasts for up to two hours[8]. These results show that microbicides containing antibodies are effective at preventing HIV in monkeys.
A similar experiment was done in 2012; it used humanized mouse models called RAG-hu mice, which contained human target cells. Results show that seven out of nine mice that were administered the VRC01 antibody and all mice that were given a cocktail containing four broadly neutralizing antibodies as a topical gel were protected against HIV-1. These results showed that broadly neutralizing antibodies could be used as a topical microbicide to prevent vaginal transmission of HIV and that a combination of antibodies can provide better protection against HIV. When the VRC01 antibody and the broadly neutralizing antibody cocktail were administered to the humanized mice via the intravenous route, none of the mice were infected with SHIV[9].
References
- ↑ 1.0 1.1 1.2 1.3 Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011 Sep 16;333(6049):1593-602. Epub 2011 Aug 11. PMID:21835983 doi:10.1126/science.1207532
- ↑ 2.0 2.1 Kwong PD, Mascola JR, Nabel GJ. The changing face of HIV vaccine research. J Int AIDS Soc. 2012 Jul 5;15(2):17407. doi: 10.7448/IAS.15.2.17407. PMID:22789610
- ↑ Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011 Sep;85(17):8954-67. Epub 2011 Jun 29. PMID:21715490 doi:10.1128/JVI.00754-11
- ↑ Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012 Jul;8(7):e1002797. Epub 2012 Jul 12. PMID:22807678 doi:10.1371/journal.ppat.1002797
- ↑ 5.0 5.1 5.2 Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010 Aug 13;329(5993):811-7. Epub 2010 Jul 8. PMID:20616231 doi:10.1126/science.1192819
- ↑ Diskin R, Scheid JF, Marcovecchio PM, West AP Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design. Science. 2011 Oct 27. PMID:22033520 doi:10.1126/science.1213782
- ↑ 7.0 7.1 Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton D, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011 Jul 14. PMID:21764753 doi:10.1126/science.1207227
- ↑ Veazey RS, Shattock RJ, Pope M, Kirijan JC, Jones J, Hu Q, Ketas T, Marx PA, Klasse PJ, Burton DR, Moore JP. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat Med. 2003 Mar;9(3):343-6. Epub 2003 Feb 10. PMID:12579198 doi:10.1038/nm833
- ↑ Veselinovic M, Neff CP, Mulder LR, Akkina R. Topical gel formulation of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 confers protection against HIV-1 vaginal challenge in a humanized mouse model. Virology. 2012 Oct 25;432(2):505-10. doi: 10.1016/j.virol.2012.06.025. Epub 2012 , Jul 24. PMID:22832125 doi:10.1016/j.virol.2012.06.025
Proteopedia Page Contributors and Editors (what is this?)
Amanda Valdiosera, Michal Harel, Chris Casey, Alexander Berchansky