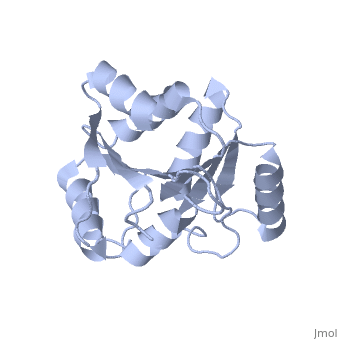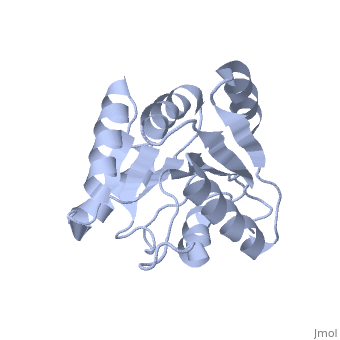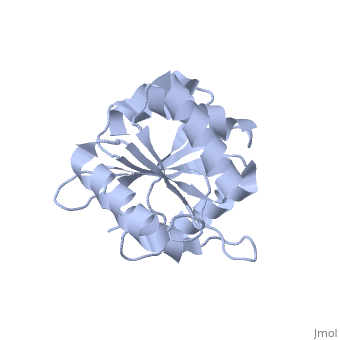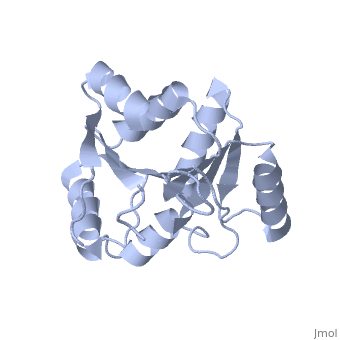
| Structural highlights
Function
[DBP5_YEAST] ATP-dependent RNA helicase associated with the nuclear pore complex and essential for mRNA export from the nucleus. May participate in a terminal step of mRNA export through the removal of proteins that accompany mRNA through the nucleopore complex. Contributes to the blocking of bulk poly(A)+ mRNA export in ethanol-stressed cells. May also be involved in early transcription.[1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11]
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
The DExD/H-box RNA-dependent ATPase Dbp5 plays an essential role in the nuclear export of mRNA. Dbp5 localizes to the nuclear pore complex, where its ATPase activity is stimulated by Gle1 and its coactivator inositol hexakisphosphate. Here, we present the crystal structure of the C-terminal domain of Dbp5, refined to 1.8 A. The structure reveals a RecA-like fold that contains two defining characteristics not present in other structurally characterized DExD/H-box proteins: a C-terminal alpha-helix and a loop connecting beta5 and alpha4, both of which are composed of conserved and unique elements in the Dbp5 primary sequence. Using structure-guided mutagenesis, we have identified several charged surface residues that, when mutated, weaken the binding of Gle1 and inhibit the ability of Gle1 to stimulate Dbp5's ATPase activity. In vivo analysis of the same mutations reveals that those mutants displaying the weakest ATPase stimulation in vitro are also unable to support yeast growth. Analysis of the correlation between the in vitro and in vivo data indicates that a threshold level of Dbp5 ATPase activity is required for cellular mRNA export that is not met by the unstimulated enzyme, suggesting a possible mechanism by which Dbp5's activity can be modulated to regulate mRNA export.
Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1.,Dossani ZY, Weirich CS, Erzberger JP, Berger JM, Weis K Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16251-6. Epub 2009 Sep 2. PMID:19805289[12]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998 May 1;17(9):2651-62. PMID:9564047 doi:http://dx.doi.org/10.1093/emboj/17.9.2651
- ↑ Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998 May 1;17(9):2663-76. PMID:9564048 doi:http://dx.doi.org/10.1093/emboj/17.9.2663
- ↑ Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999 Aug 2;18(15):4332-47. PMID:10428971 doi:http://dx.doi.org/10.1093/emboj/18.15.4332
- ↑ Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 1999 Oct 15;18(20):5761-77. PMID:10610322 doi:10.1093/emboj/18.20.5761
- ↑ Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999 Oct 15;18(20):5778-88. PMID:10523319 doi:10.1093/emboj/18.20.5778
- ↑ Hilleren P, Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3'-end formation of nascent transcripts. RNA. 2001 May;7(5):753-64. PMID:11350039
- ↑ Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3' processing, and mRNA export. Mol Cell Biol. 2002 Sep;22(18):6441-57. PMID:12192043
- ↑ Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell. 2003 Apr;14(4):1664-76. PMID:12686617 doi:http://dx.doi.org/10.1091/mbc.E02-09-0602
- ↑ Takemura R, Inoue Y, Izawa S. Stress response in yeast mRNA export factor: reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J Cell Sci. 2004 Aug 15;117(Pt 18):4189-97. Epub 2004 Jul 27. PMID:15280434 doi:http://dx.doi.org/10.1242/jcs.01296
- ↑ Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell. 2004 Dec 3;16(5):749-60. PMID:15574330 doi:10.1016/j.molcel.2004.10.032
- ↑ Estruch F, Hodge CA, Rodriguez-Navarro S, Cole CN. Physical and genetic interactions link the yeast protein Zds1p with mRNA nuclear export. J Biol Chem. 2005 Mar 11;280(10):9691-7. Epub 2004 Dec 24. PMID:15619606 doi:http://dx.doi.org/M413025200
- ↑ Dossani ZY, Weirich CS, Erzberger JP, Berger JM, Weis K. Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16251-6. Epub 2009 Sep 2. PMID:19805289
|