2hym
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:2hym.gif|left|200px]] | + | [[Image:2hym.gif|left|200px]] |
| - | + | ||
| - | '''NMR based Docking Model of the Complex between the Human Type I Interferon Receptor and Human Interferon alpha-2''' | + | {{Structure |
| + | |PDB= 2hym |SIZE=350|CAPTION= <scene name='initialview01'>2hym</scene> | ||
| + | |SITE= | ||
| + | |LIGAND= | ||
| + | |ACTIVITY= | ||
| + | |GENE= IFNABR ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]), IFNA2 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]) | ||
| + | }} | ||
| + | |||
| + | '''NMR based Docking Model of the Complex between the Human Type I Interferon Receptor and Human Interferon alpha-2''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
| - | 2HYM is a [ | + | 2HYM is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2HYM OCA]. |
==Reference== | ==Reference== | ||
| - | Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex., Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J, Protein Sci. 2006 Nov;15(11):2656-68. Epub 2006 Sep 25. PMID:[http:// | + | Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex., Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J, Protein Sci. 2006 Nov;15(11):2656-68. Epub 2006 Sep 25. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17001036 17001036] |
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
| Line 20: | Line 29: | ||
[[Category: interferon receptor complex]] | [[Category: interferon receptor complex]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 17:23:13 2008'' |
Revision as of 15:23, 20 March 2008
| |||||||
| Gene: | IFNABR (Homo sapiens), IFNA2 (Homo sapiens) | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
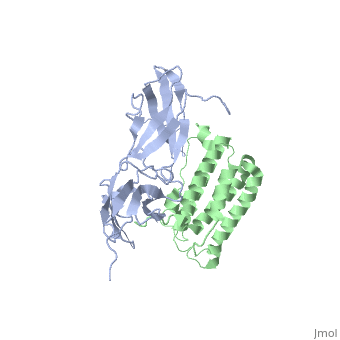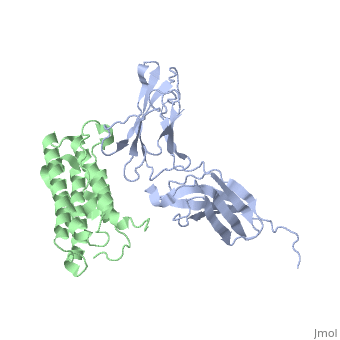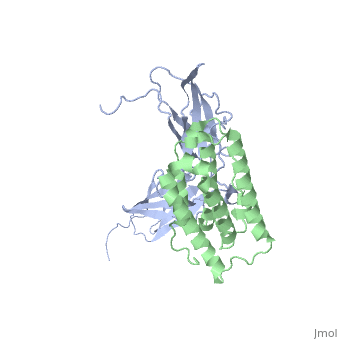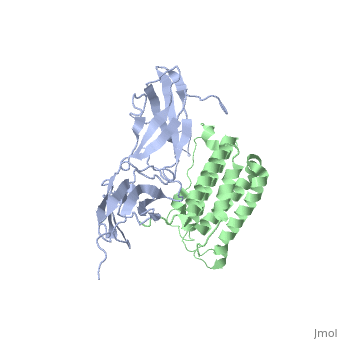
NMR based Docking Model of the Complex between the Human Type I Interferon Receptor and Human Interferon alpha-2
Overview
Type I interferons (IFNs) are a family of homologous helical cytokines that exhibit pleiotropic effects on a wide variety of cell types, including antiviral activity and antibacterial, antiprozoal, immunomodulatory, and cell growth regulatory functions. Consequently, IFNs are the human proteins most widely used in the treatment of several kinds of cancer, hepatitis C, and multiple sclerosis. All type I IFNs bind to a cell surface receptor consisting of two subunits, IFNAR1 and IFNAR2, associating upon binding of interferon. The structure of the extracellular domain of IFNAR2 (R2-EC) was solved recently. Here we study the complex and the binding interface of IFNalpha2 with R2-EC using multidimensional NMR techniques. NMR shows that IFNalpha2 does not undergo significant structural changes upon binding to its receptor, suggesting a lock-and-key mechanism for binding. Cross saturation experiments were used to determine the receptor binding site upon IFNalpha2. The NMR data and previously published mutagenesis data were used to derive a docking model of the complex with an RMSD of 1 Angstrom, and its well-defined orientation between IFNalpha2 and R2-EC and the structural quality greatly improve upon previously suggested models. The relative ligand-receptor orientation is believed to be important for interferon signaling and possibly one of the parameters that distinguish the different IFN I subtypes. This structural information provides important insight into interferon signaling processes and may allow improvement in the development of therapeutically used IFNs and IFN-like molecules.
About this Structure
2HYM is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex., Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J, Protein Sci. 2006 Nov;15(11):2656-68. Epub 2006 Sep 25. PMID:17001036
Page seeded by OCA on Thu Mar 20 17:23:13 2008