We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 967
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
==Structure of the Mouse RNase H2 Complex== | ==Structure of the Mouse RNase H2 Complex== | ||
<StructureSection load='3kio' size='400' side='right' caption='mouse RNase H2 complex, (PDB code [[3kio]])> | <StructureSection load='3kio' size='400' side='right' caption='mouse RNase H2 complex, (PDB code [[3kio]])> | ||
| - | '''The RNase H2 ribonuclease complex''' is a heterotrimeric endoribonuclease responsible for the major ribonuclease H activity in mammalian cells. In mouse, the complex is encoded by 3 genes located on chromosomes 8 (''Rnaseh2a''), 14 (''Rnaseh2b'') and 19 (''Rnaseh2c''). This enzyme specifically cleaves the 3’O-Phosphate bond of RNA in a DNA/RNA hybrids to produce 5’ phosphate and 3’hydroxyl ends. | + | '''The RNase H2 ribonuclease complex''' is a heterotrimeric endoribonuclease responsible for the major ribonuclease H activity in mammalian cells. In mouse, the complex is encoded by 3 genes located on chromosomes 8 (''Rnaseh2a''), 14 (''Rnaseh2b'') and 19 (''Rnaseh2c'')<ref> http://genome-euro.ucsc.edu/cgi-bin/hgTracks?clade=mammal&org=Mouse&db=mm10&position=RnaseH2&hgt.positionInput=RnaseH2&hgt.suggestTrack=knownGene&Submit=submit&hgsid=201143152_yP1Xd4bMnHS7DV0d3VcqpDSxzzuQ&pix=1563</ref>. This enzyme specifically cleaves the 3’O-Phosphate bond of RNA in a DNA/RNA hybrids to produce 5’ phosphate and 3’hydroxyl ends. |
== Biological role == | == Biological role == | ||
Ribonucleases H are the only known enzymes, able to degrade the RNA strand of a DNA/RNA hybrid in a sequence-nonspecific way. | Ribonucleases H are the only known enzymes, able to degrade the RNA strand of a DNA/RNA hybrid in a sequence-nonspecific way. | ||
| - | There are two types of RNase H (RNases H1 and RNases H2) classified according to their sequence conservation and substrate preference. Currently, three types of RNA/DNA hybrids are known: simple RNA/DNA duplexes ('''Figure 1A'''), RNA•DNA/DNA hybrids ('''Figure 1B'''), and DNA•RNAfew•DNA/DNA hybrids ('''Figure 1C'''). RNases H2 is totally able to cleave a single ribonucleotide embedded in a double strand DNA (DNA• RNAfew •DNA/DNA type) when RNases H1 require at least 4 ribonucleotides. This ability and their high expression in proliferating cells suggest that RNases H2 are involved in DNA repair and replication. | + | There are two types of RNase H (RNases H1 and RNases H2) classified according to their sequence conservation and substrate preference. Currently, three types of RNA/DNA hybrids are known: simple RNA/DNA duplexes ('''Figure 1A'''), RNA•DNA/DNA hybrids ('''Figure 1B'''), and DNA•RNAfew•DNA/DNA hybrids ('''Figure 1C'''). RNases H2 is totally able to cleave a single ribonucleotide embedded in a double strand DNA (DNA• RNAfew •DNA/DNA type) when RNases H1 require at least 4 ribonucleotides. This ability and their high expression in proliferating cells suggest that RNases H2 are involved in DNA repair and replication<ref> Rychlik, Monika P., Hyongi Chon, Susana M. Cerritelli, Paulina Klimek, Robert J. Crouch, and Marcin Nowotny. “Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.” Molecular Cell 40, no. 4 (November 24, 2010): 658–70. doi:10.1016/j.molcel.2010.11.001.</ref>. |
[[Image:ThreetypesofRNA-DNAhybrids.png|300px|left|thumb| '''Figure 1''' : Three types of RNA/DNA hybrids]] | [[Image:ThreetypesofRNA-DNAhybrids.png|300px|left|thumb| '''Figure 1''' : Three types of RNA/DNA hybrids]] | ||
| - | Indeed, ribonucleotides are wrongly incorporated into DNA during DNA replication at a frequency of about 2 ribonucleotides per kb. With such frequency, these errors are by far the most abundant threat of DNA damaging. Hence, a correction is essential to the preservation of DNA integrity: the most common correction mechanism involves RNases H2 and is called Ribonucleotide Excision Repair (RER). The incorporation of ribonucleotides in DNA produce DNA•RNAfew•DNA/DNA hybrids from which the few misincorporated ribonucleotides can be removed by an RNase H2. | + | Indeed, ribonucleotides are wrongly incorporated into DNA during DNA replication at a frequency of about 2 ribonucleotides per kb. With such frequency, these errors are by far the most abundant threat of DNA damaging. Hence, a correction is essential to the preservation of DNA integrity: the most common correction mechanism involves RNases H2 and is called Ribonucleotide Excision Repair (RER). The incorporation of ribonucleotides in DNA produce DNA•RNAfew•DNA/DNA hybrids from which the few misincorporated ribonucleotides can be removed by an RNase H2<ref> Sparks, Justin L., Hyongi Chon, Susana M. Cerritelli, Thomas A. Kunkel, Erik Johansson, Robert J. Crouch, and Peter M. Burgers. “RNase H2-Initiated Ribonucleotide Excision Repair.” Molecular Cell 47, no. 6 (September 28, 2012): 980–86. doi:10.1016/j.molcel.2012.06.035.</ref>. |
| - | This repair activity is guided by the interaction between C-terminus of RNase H2B protein and the DNA clamp PCNA. This interaction occurs through a hydrophobic conserved peptide motif called the PCNA interaction peptide PIP (PIP-box: Residues 294 to 301 MKSIDTFF of H2B protein) that interacts with a hydrophobic groove near the PCNA C-terminus. This interaction allows RNase H2 to scan DNA for misincorporated ribonucleotides which makes the Ribonucleotide Excision Repair more efficient. | + | This repair activity is guided by the interaction between C-terminus of RNase H2B protein and the DNA clamp PCNA. This interaction occurs through a hydrophobic conserved peptide motif called the PCNA interaction peptide PIP (PIP-box: Residues 294 to 301 MKSIDTFF of H2B protein) that interacts with a hydrophobic groove near the PCNA C-terminus. This interaction allows RNase H2 to scan DNA for misincorporated ribonucleotides which makes the Ribonucleotide Excision Repair more efficient<ref> Bubeck, Doryen, Martin A. M. Reijns, Stephen C. Graham, Katy R. Astell, E. Yvonne Jones, and Andrew P. Jackson. “PCNA Directs Type 2 RNase H Activity on DNA Replication and Repair Substrates.” Nucleic Acids Research 39, no. 9 (May 2011): 3652–66. [http://dx.doi.org/10.1093/nar/gkq980 doi:10.1093/nar/gkq980.</ref>. |
| - | + | ||
Furthermore, ''in vitro'' studies have shown that RNases H2 is likely to be involved in the removal of RNA primer from Okazaki fragment produced during the synthesis of the lagging strand in DNA replication since Okazaki fragment are RNA•DNA/DNA hybrids ('''Figure 1B'''). | Furthermore, ''in vitro'' studies have shown that RNases H2 is likely to be involved in the removal of RNA primer from Okazaki fragment produced during the synthesis of the lagging strand in DNA replication since Okazaki fragment are RNA•DNA/DNA hybrids ('''Figure 1B'''). | ||
| Line 33: | Line 32: | ||
H2C protein is found in the middle of the elongated complex structure, flanked by H2A and H2B proteins on the ends. | H2C protein is found in the middle of the elongated complex structure, flanked by H2A and H2B proteins on the ends. | ||
| - | The complex is stabilized by the intimately interwoven architecture of H2B and H2C: The N-terminal region of H2B protein (amino acids 1-92) weaves together with H2C domain to form 3 β-barrels, also called | + | The complex is stabilized by the intimately interwoven architecture of H2B and H2C: The N-terminal region of H2B protein (amino acids 1-92) weaves together with H2C domain to form 3 β-barrels, also called “triple barrel”. This triple barrel is formed from a total of 18 β-sheets and produces a pseudo-2-fold axis of symmetry along the central barrel. Also, it permits to leave the mostly α-helical C-terminal region of H2B available for potential interactions with other protein (for example the PCNA protein). Finally, it has been found that the motif provides a platform for securely binding the H2A protein: the side and end of the first barrel in the subcomplex H2B/H2C form a tight interface with amino acids 197-258 in the C-terminal region of H2A protein. This interface is composed mainly of hydrophobic residues. |
| Line 48: | Line 47: | ||
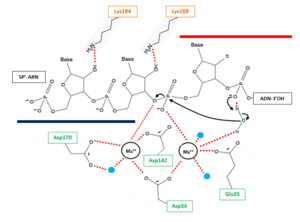
The RNase H2 recognize 2’OH group of ribonucleotides in RNA at RNA/DNA junction and cannot cleave unhybridized RNA. The phosphodiester hydrolysis catalysed by the RNase H2 is likely following a two-metal ion-dependent mechanism quite common for phosphoryl hydrolases like RNase H enzymes. | The RNase H2 recognize 2’OH group of ribonucleotides in RNA at RNA/DNA junction and cannot cleave unhybridized RNA. The phosphodiester hydrolysis catalysed by the RNase H2 is likely following a two-metal ion-dependent mechanism quite common for phosphoryl hydrolases like RNase H enzymes. | ||
| - | The active site of the catalytic H2A protein is located in a cleft near one end of the complex and formed by strands β2 and | + | The active site of the catalytic H2A protein is located in a cleft near one end of the complex and formed by strands β2 and 5, and helices α4 and α5. The active site contains four catalytic amino acids strictly conserved in RNases H2: <scene name='60/604486/Site_actif/1'>Asp34, Glu35, Asp142 and Asp170</scene>. These amino acids are essential for the coordination of the 2 divalent metal ions implicated in the stabilisation of reaction intermediates and transition states. In vitro, these ions can be Mg++, Mn++ or Zn++ but the native enzyme is likely to contain Mg++. |
[[Image:ProposedmechanismforOkazaki.png|300px|left|thumb| '''Figure 2''' : Proposed mechanism for Okazaki fragment processing (not a concerted mechanism)]] | [[Image:ProposedmechanismforOkazaki.png|300px|left|thumb| '''Figure 2''' : Proposed mechanism for Okazaki fragment processing (not a concerted mechanism)]] | ||
Revision as of 16:38, 8 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
Structure of the Mouse RNase H2 Complex
| |||||||||||
References
- ↑ http://genome-euro.ucsc.edu/cgi-bin/hgTracks?clade=mammal&org=Mouse&db=mm10&position=RnaseH2&hgt.positionInput=RnaseH2&hgt.suggestTrack=knownGene&Submit=submit&hgsid=201143152_yP1Xd4bMnHS7DV0d3VcqpDSxzzuQ&pix=1563
- ↑ Rychlik, Monika P., Hyongi Chon, Susana M. Cerritelli, Paulina Klimek, Robert J. Crouch, and Marcin Nowotny. “Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.” Molecular Cell 40, no. 4 (November 24, 2010): 658–70. doi:10.1016/j.molcel.2010.11.001.
- ↑ Sparks, Justin L., Hyongi Chon, Susana M. Cerritelli, Thomas A. Kunkel, Erik Johansson, Robert J. Crouch, and Peter M. Burgers. “RNase H2-Initiated Ribonucleotide Excision Repair.” Molecular Cell 47, no. 6 (September 28, 2012): 980–86. doi:10.1016/j.molcel.2012.06.035.
- ↑ Bubeck, Doryen, Martin A. M. Reijns, Stephen C. Graham, Katy R. Astell, E. Yvonne Jones, and Andrew P. Jackson. “PCNA Directs Type 2 RNase H Activity on DNA Replication and Repair Substrates.” Nucleic Acids Research 39, no. 9 (May 2011): 3652–66. [http://dx.doi.org/10.1093/nar/gkq980 doi:10.1093/nar/gkq980.