Magainin 2
From Proteopedia
| Line 17: | Line 17: | ||
== Structural highlights and antimicrobial mechanism == | == Structural highlights and antimicrobial mechanism == | ||
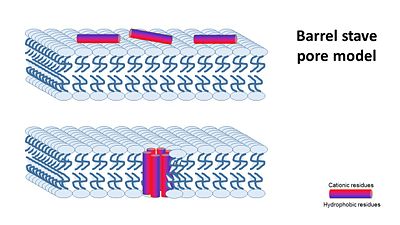
In general, amphipathic helical peptides that disrupt the ionic gradient of cells are thought to do so in various suggested mechanisms. One of the models is called the Barrel stave pore model. In this suggested mechanism peptides bind to the cell membrane, and in the seconed step the form ion channels assembled from 4–6 peptide molecules in the bacterial membrane. This results in the insides of the cell leaking outside, causing cell death. | In general, amphipathic helical peptides that disrupt the ionic gradient of cells are thought to do so in various suggested mechanisms. One of the models is called the Barrel stave pore model. In this suggested mechanism peptides bind to the cell membrane, and in the seconed step the form ion channels assembled from 4–6 peptide molecules in the bacterial membrane. This results in the insides of the cell leaking outside, causing cell death. | ||
| - | [[Image:Barrel_stave_pore_model.JPG]] | + | [[Image:Barrel_stave_pore_model.JPG|center|400px]] |
| - | (Image is according to Wimley, 2010. | + | (Image is according to Wimley, 2010.) |
It was thought that this mechanism is also acountable for Magainin 2, But earlier solid-state NMR results show that its helix axis lies in the plane of phospholipid bilayers, suggesting that magainin’s mechanism for disrupting the ionic gradient may be fundamentally different. Therefore it's mechanism is still unclear, but either way it seems like Magainin 2 binds to the bacterial membrane to cause it's antibacterial effect. | It was thought that this mechanism is also acountable for Magainin 2, But earlier solid-state NMR results show that its helix axis lies in the plane of phospholipid bilayers, suggesting that magainin’s mechanism for disrupting the ionic gradient may be fundamentally different. Therefore it's mechanism is still unclear, but either way it seems like Magainin 2 binds to the bacterial membrane to cause it's antibacterial effect. | ||
If we look at the Magainin 2 structure we can see how it allows Magainin 2 to bind to membranes: | If we look at the Magainin 2 structure we can see how it allows Magainin 2 to bind to membranes: | ||
Revision as of 15:00, 1 February 2015
Magainin 2
| |||||||||||
References
1-J. Gesella., M. Zasloffb and S. J. Opellaa., Two-dimensional H NMR experiments show that the 23-residue magainin antibiotic peptide is an α-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. Journal of Biomolecular NMR, 1997. 9: 127–135.
2-Z. Hayouka., D. E. Mortenson., D. F. Kreitler., B. Weisblum., K. T. Forest, and S. H. Gellman., Evidence for Phenylalanine Zipper-Mediated Dimerization in the X‑ray Crystal Structure of a Magainin 2 Analogue. J. Am. Chem. Soc, 2013. 135: 15738−15741.
3- Y. Tamba & M. Yamazaki.,Magainin 2-Induced Pore Formation in the Lipid Membranes Depends on Its Concentration in the Membrane Interface. J. Phys. Chem. B, 2009. 113: 4846–4852.
4- W. C. Wimley., Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS CHEMICAL BIOLOGY, 2010. 10: 905-917.