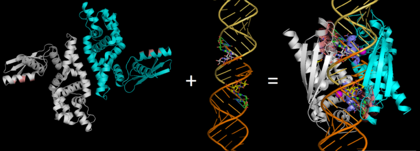
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1085
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
[[Image: Cleavage site of RNase III.png|thumb|right|320px|Fig.3. ''cleavage site of Aa-RNase III'' ]] | [[Image: Cleavage site of RNase III.png|thumb|right|320px|Fig.3. ''cleavage site of Aa-RNase III'' ]] | ||
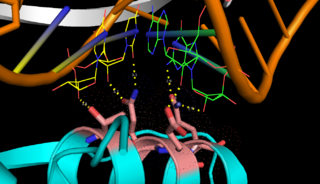
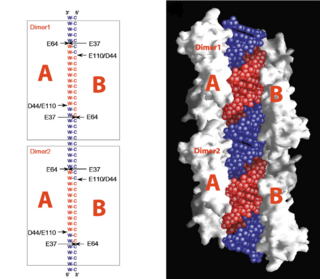
| - | On the basis of the structural and biochemical data, catalytic models were proposed before the structure of a catalytic complex became available. The crystal structure shows that Aa-RNase III is composed of a <scene name='69/699998/Dimer/3'>symmetric dimer</scene>. In addition, in vivo data suggested that E110, E37, D44, and E64 are essential for catalysis<ref>Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466.</ref>. This led to the model of the proteins active centers, which can accommodate a dsRNA substrate, each containing two different RNA cleavage sites, <scene name='69/699998/D44_e110_e37_e64/1'>D44/E110 and E37/E64</scene> (pink and red). Specifically, E64 from each partner subunit, along with E37, E40, and D44 are located in the signature motif located at each end of the catalytic valley<ref>Blaszczyk, J., et al., Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure, 2001. 9(12): p. 1225-1236</ref>. Comparing the structure of Aa-RNase III with the structure of RNA-free Thermotoga maritima RNase III (RNA-free Tm-RNase III, PDB ID code 1O0W)<ref>Gan, J., et al., Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure, 2005. 13(10): p. 1435-1442.</ref> shows that there is <scene name='69/699998/Linker_rotation/1'>dramatic rotation</scene> and shift of dsRBD due to RNA binding, and there is a <scene name='69/699998/Linker/2'>flexible linker</scene> KGEMLFD between endoRD and dsRBD leading the rotation happend. In addtion, the two dsRBDs are apart from each other, allowing free rotation of dsRBDdsRNA around the linker. | + | On the basis of the structural and biochemical data, catalytic models were proposed before the structure of a catalytic complex became available. The crystal structure shows that Aa-RNase III is composed of a <scene name='69/699998/Dimer/3'>symmetric dimer</scene>. In addition, in vivo data suggested that E110, E37, D44, and E64 are essential for catalysis<ref>Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466.</ref>. This led to the model of the proteins active centers, which can accommodate a dsRNA substrate, each containing two different RNA cleavage sites, <scene name='69/699998/D44_e110_e37_e64/1'>D44/E110 and E37/E64</scene> (pink and red). Specifically, E64 from each partner subunit, along with E37, E40, and D44 are located in the signature motif located at each end of the catalytic valley<ref>Blaszczyk, J., et al., Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure, 2001. 9(12): p. 1225-1236</ref>. Comparing the structure of Aa-RNase III with the structure of RNA-free Thermotoga maritima RNase III (RNA-free Tm-RNase III, PDB ID code 1O0W)<ref name= Gan>Gan, J., et al., Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure, 2005. 13(10): p. 1435-1442.</ref> shows that there is <scene name='69/699998/Linker_rotation/1'>dramatic rotation</scene> and shift of dsRBD due to RNA binding, and there is a <scene name='69/699998/Linker/2'>flexible linker</scene> KGEMLFD between endoRD and dsRBD leading the rotation happend. In addtion, the two dsRBDs are apart from each other, allowing free rotation of dsRBDdsRNA around the linker. |
| Line 21: | Line 21: | ||
[[Image: Ge470405.f7.gif|thumb|left|640px|Fig.4. ''Hypothetical Pathway Leading to the two Functional Forms of RNase III'' ]] | [[Image: Ge470405.f7.gif|thumb|left|640px|Fig.4. ''Hypothetical Pathway Leading to the two Functional Forms of RNase III'' ]] | ||
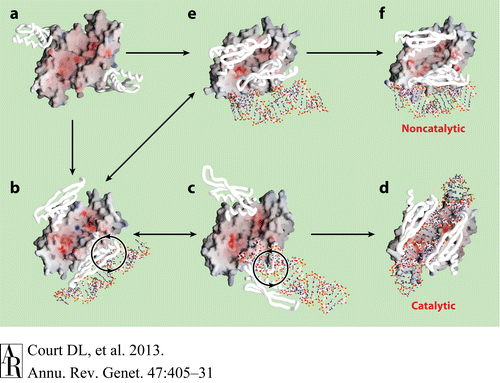
| - | Catalytic processes in addition to the catalytic and noncatalytic forms of the Aa-RNase III–dsRNA structures revealed possible intermediate states of dsRNA binding and a hypothetical pathway of RNase III in vivo. The RNA-free RNase III dimer in conformation A (Figure a, PDB 1O0W) first binds a dsRNA with one of the two dsRBDs. The resulting complex may have at least two possible conformations, B (Figure b, PDB1YZ9 AaRNase IIIE110Q) and E (Figure e, PDB 1YYO), in which the dsRNA is located outside of the catalytic valley. These two conformations, b and e, appear to be interchangeable. In conformation E, the two dsRBDs pack against each other, hindering free rotation of the dsRBD-dsRNA complex around the flexible linker between the endoND and dsRBD. Conformation b allows free rotation of dsRBD-dsRNA around the linker (Figure b). The clockwise rotation of dsRBD-dsRNA leads to the catalytic form (Figure d, PDB 2EZ6) via conformation C (Figure c, PDB 1YYW). Conformations B and C are also likely to be interchangeable. In contrast, a factor that uncouples binding and processing can further stabilize conformation E leading to the noncatalytic form of RNase III (Figure f, PDB 1RC7)[<ref>Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466</ref>, <ref>Gan, J., et al., Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell, 2006. 124(2): p. 355-66</ref>, <ref | + | Catalytic processes in addition to the catalytic and noncatalytic forms of the Aa-RNase III–dsRNA structures revealed possible intermediate states of dsRNA binding and a hypothetical pathway of RNase III in vivo. The RNA-free RNase III dimer in conformation A (Figure a, PDB 1O0W) first binds a dsRNA with one of the two dsRBDs. The resulting complex may have at least two possible conformations, B (Figure b, PDB1YZ9 AaRNase IIIE110Q) and E (Figure e, PDB 1YYO), in which the dsRNA is located outside of the catalytic valley. These two conformations, b and e, appear to be interchangeable. In conformation E, the two dsRBDs pack against each other, hindering free rotation of the dsRBD-dsRNA complex around the flexible linker between the endoND and dsRBD. Conformation b allows free rotation of dsRBD-dsRNA around the linker (Figure b). The clockwise rotation of dsRBD-dsRNA leads to the catalytic form (Figure d, PDB 2EZ6) via conformation C (Figure c, PDB 1YYW). Conformations B and C are also likely to be interchangeable. In contrast, a factor that uncouples binding and processing can further stabilize conformation E leading to the noncatalytic form of RNase III (Figure f, PDB 1RC7)[<ref>Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466</ref>, <ref>Gan, J., et al., Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell, 2006. 124(2): p. 355-66</ref>, <ref name =Gan/>]. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 12:55, 23 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Endoribonuclease III
| |||||||||||
References
- ↑ Lioliou E, Sharma CM, Caldelari I, et al. Global Regulatory Functions of the Staphylococcus aureus Endoribonuclease III in Gene Expression. Hughes D, ed. PLoS Genetics. 2012;8(6):e1002782. doi:10.1371/journal.pgen.1002782
- ↑ Lamontagne, B., et al., The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Yeast, 2001. 45(191): p. 154-158.
- ↑ Robertson, H.D., Escherichia coli ribonuclease III cleavage sites. Cell, 1982. 30(3): p. 669-672.
- ↑ Grunberg-Manago, M., Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annual review of genetics, 1999. 33(1): p. 193-227.
- ↑ Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466.
- ↑ Blaszczyk, J., et al., Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure, 2001. 9(12): p. 1225-1236
- ↑ 7.0 7.1 Gan, J., et al., Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure, 2005. 13(10): p. 1435-1442.
- ↑ Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466
- ↑ Gan, J., et al., Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell, 2006. 124(2): p. 355-66