We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1085
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Exploring the Structure == | == Exploring the Structure == | ||
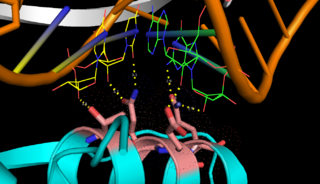
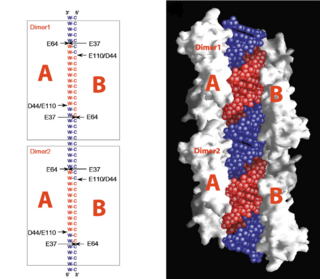
| - | <scene name='69/699998/Monomer/3'>Monomers</scene> of Aquifex aeolicus RNase III (Aa-RNase III) are composed of an <scene name='69/699998/Endond/2'>endonuclease domain</scene> (endoND, green) and a <scene name='69/699998/Dsrbd/4'>dsRNA binding domain</scene> (dsRBD, red)<ref>Lamontagne, B., et al., The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Yeast, 2001. 45(191): p. 154-158.</ref>. The crystal structure shows that RNase III is composed of a <scene name='69/699998/Dimer/4'>symmetric dimer</scene>. The sequence of the endoND is characterized by a stretch of conserved residues (37ERLEFLGD44 in Aa-RNase III), which is known as the RNase III <scene name='69/699998/Sig_motiff/2'>signature motif</scene>(purple) and makes up a large part of the active center, where catalysis takes place. RNase III hydrogen-bonds to dsRNA with residues <scene name='69/699998/Bindingsite/1'>T154,Q157,E158,Q161</scene>, which make up the binding site (''Fig.1''). RNase III can affect gene expression in either of two ways: as a processing enzyme | + | <scene name='69/699998/Monomer/3'>Monomers</scene> of Aquifex aeolicus RNase III (Aa-RNase III) are composed of an <scene name='69/699998/Endond/2'>endonuclease domain</scene> (endoND, green) and a <scene name='69/699998/Dsrbd/4'>dsRNA binding domain</scene> (dsRBD, red)<ref>Lamontagne, B., et al., The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Yeast, 2001. 45(191): p. 154-158.</ref>. The crystal structure shows that RNase III is composed of a <scene name='69/699998/Dimer/4'>symmetric dimer</scene>. The sequence of the endoND is characterized by a stretch of conserved residues (37ERLEFLGD44 in Aa-RNase III), which is known as the RNase III <scene name='69/699998/Sig_motiff/2'>signature motif</scene>(purple) and makes up a large part of the active center, where catalysis takes place. RNase III hydrogen-bonds to dsRNA with residues <scene name='69/699998/Bindingsite/1'>T154,Q157,E158,Q161</scene>, which make up the binding site (''Fig.1''). RNase III can affect gene expression in either of two ways: as a processing enzyme, where RNase III cleaves both natural and synthetic dsRNA into small duplex products averaging 9–18 base pairs in length, or as a binding protein which binds and stabilizes certain RNAs, thus suppressing the expression of certain genes[<ref>Robertson, H.D., Escherichia coli ribonuclease III cleavage sites. Cell, 1982. 30(3): p. 669-672.</ref>, <ref>Grunberg-Manago, M., Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annual review of genetics, 1999. 33(1): p. 193-227.</ref>]. |
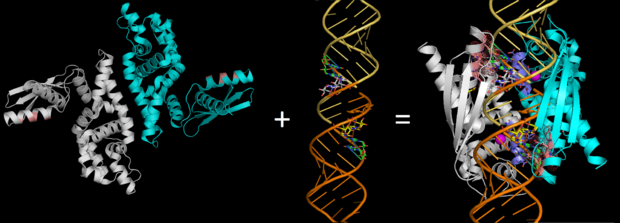
[[Image: ALIGN2.png|thumb|left|620px|Fig.2. ''RNase III (Pbd codes 1O0W & 2EZ6) conformation change when bound to dsRNA'' ]] | [[Image: ALIGN2.png|thumb|left|620px|Fig.2. ''RNase III (Pbd codes 1O0W & 2EZ6) conformation change when bound to dsRNA'' ]] | ||
Revision as of 16:02, 23 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Endoribonuclease III
| |||||||||||
References
- ↑ Lioliou E, Sharma CM, Caldelari I, et al. Global Regulatory Functions of the Staphylococcus aureus Endoribonuclease III in Gene Expression. Hughes D, ed. PLoS Genetics. 2012;8(6):e1002782. doi:10.1371/journal.pgen.1002782
- ↑ Lamontagne, B., et al., The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Yeast, 2001. 45(191): p. 154-158.
- ↑ Robertson, H.D., Escherichia coli ribonuclease III cleavage sites. Cell, 1982. 30(3): p. 669-672.
- ↑ Grunberg-Manago, M., Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annual review of genetics, 1999. 33(1): p. 193-227.
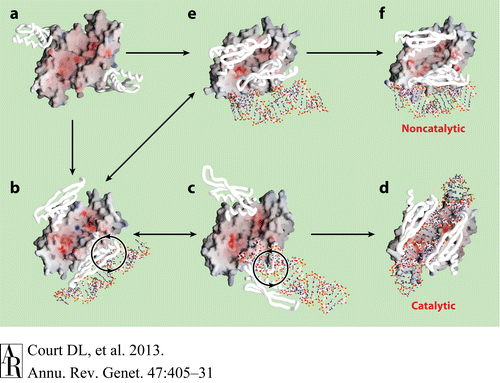
- ↑ 5.0 5.1 Gan, J., et al., Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure, 2005. 13(10): p. 1435-1442.
- ↑ Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466.
- ↑ Blaszczyk, J., et al., Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure, 2001. 9(12): p. 1225-1236
- ↑ Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466
- ↑ Gan, J., et al., Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell, 2006. 124(2): p. 355-66