We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1085
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
Universally, it has been shown to mediate RNA turnover at the post-transcriptional level through processing rRNAs, tRNAs, some mRNAs, as well as non-coding dsRNAs. In microbes, RNase III has been shown that it also represses the synthesis of virulence factors (through the cleavage of foreign RNA.) <ref>Lioliou E, Sharma CM, Caldelari I, et al. Global Regulatory Functions of the Staphylococcus aureus Endoribonuclease III in Gene Expression. Hughes D, ed. PLoS Genetics. 2012;8(6):e1002782. doi:10.1371/journal.pgen.1002782</ref> | Universally, it has been shown to mediate RNA turnover at the post-transcriptional level through processing rRNAs, tRNAs, some mRNAs, as well as non-coding dsRNAs. In microbes, RNase III has been shown that it also represses the synthesis of virulence factors (through the cleavage of foreign RNA.) <ref>Lioliou E, Sharma CM, Caldelari I, et al. Global Regulatory Functions of the Staphylococcus aureus Endoribonuclease III in Gene Expression. Hughes D, ed. PLoS Genetics. 2012;8(6):e1002782. doi:10.1371/journal.pgen.1002782</ref> | ||
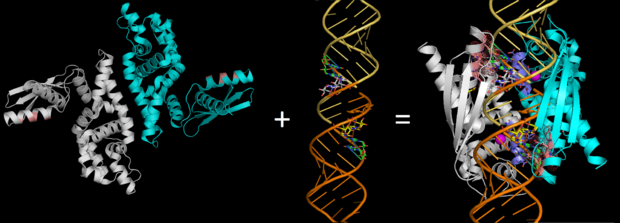
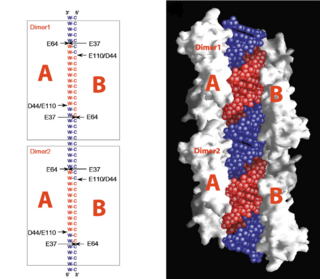
| - | In eukaryotes, RNase III has also been shown to generate microRNAs and siRNAs, which are central to gene regulation pathways. Model organisms for RNase III include E. coli and Aquifex aeolicus, whose structures have been solved with x-ray crystallography. | + | In eukaryotes, RNase III has also been shown to generate microRNAs and siRNAs, which are central to gene regulation pathways. Model organisms for RNase III include E. coli and ''Aquifex aeolicus'', whose structures have been solved with x-ray crystallography. Below, the structures of RNase III from ''Aquifex aeolicus'' and ''Thermotoga maritima'' are used to study the structure and correlating function in more detail. These organisms RNase IIIs have been compared previously due to their structural similarity. |
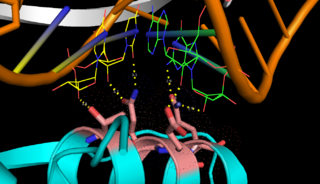
[[Image: Binding site 1.png|thumb|left|320px|Fig.1. ''RNase III binding to dsRNA'' ]] | [[Image: Binding site 1.png|thumb|left|320px|Fig.1. ''RNase III binding to dsRNA'' ]] | ||
Revision as of 05:40, 24 April 2015
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Endoribonuclease III
| |||||||||||
References
- ↑ Lioliou E, Sharma CM, Caldelari I, et al. Global Regulatory Functions of the Staphylococcus aureus Endoribonuclease III in Gene Expression. Hughes D, ed. PLoS Genetics. 2012;8(6):e1002782. doi:10.1371/journal.pgen.1002782
- ↑ Lamontagne, B., et al., The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Yeast, 2001. 45(191): p. 154-158.
- ↑ Robertson, H.D., Escherichia coli ribonuclease III cleavage sites. Cell, 1982. 30(3): p. 669-672.
- ↑ Grunberg-Manago, M., Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annual review of genetics, 1999. 33(1): p. 193-227.
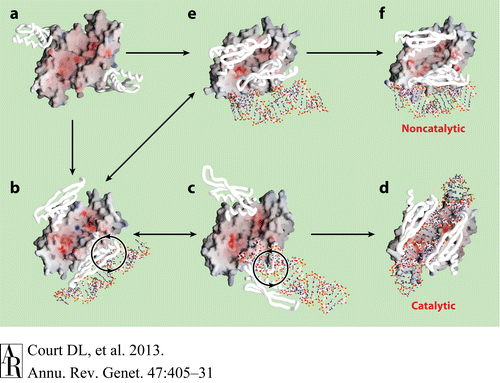
- ↑ 5.0 5.1 Gan, J., et al., Intermediate states of ribonuclease III in complex with double-stranded RNA. Structure, 2005. 13(10): p. 1435-1442.
- ↑ Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466.
- ↑ Blaszczyk, J., et al., Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure, 2001. 9(12): p. 1225-1236
- ↑ Blaszczyk, J., et al., Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure, 2004. 12(3): p. 457-466
- ↑ Gan, J., et al., Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell, 2006. 124(2): p. 355-66