User:Jonathan Lloyd/Sandbox 1
From Proteopedia
| Line 1: | Line 1: | ||
==ATAD2b== | ==ATAD2b== | ||
| - | <StructureSection load='ATAD2b.pdb' size='340' side='right' caption='Cartoon showing the bromodomain structure of human ATAD2b. The ATAD2b bromodamain has a conserved structural fold of a left-handed bundle of four helices (αZ, αA, αB, and αC ). This Cartoon was generated by using PyMol' scene=''> | + | <StructureSection load='ATAD2b.pdb' size='340' side='right' caption='Figure 2. Cartoon showing the bromodomain structure of human ATAD2b. The ATAD2b bromodamain has a conserved structural fold of a left-handed bundle of four helices (αZ, αA, αB, and αC ). This Cartoon was generated by using PyMol' scene=''> |
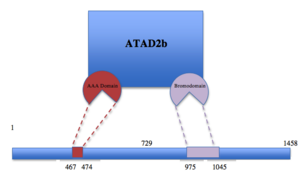
| - | [[Image:table 5.png|thumb|300px|left|Schematic representation of ATAD2b with the two different subunits shown in different colors.]] | + | [[Image:table 5.png|thumb|300px|left|Figure 1. Schematic representation of ATAD2b with the two different subunits shown in different colors.]] |
ATAD2b (also known as KIAA1240) is an eukaryotic protein located in the nucleus that contains an AAA domain and a bromodomain. AAA ATPase domains use the energy of adenosine tri-phosphate (ATP) binding to participate in cellular activities as diverse as cell cycle control, signal transduction, disassembly of macromolecular complexes and regulation of gene expression. Bromodomains bind acetyl-lysine motifs and are thought to regulate protein–protein interactions in chromatin remodeling and transcriptional control. In humans, ATAD2B is an E2F target gene that binds to the MYC oncogene. High ATAD2b levels correlate with a higher risk of distant recurrence in breast cancer, and mutation of the bromodomain in ATAD2b impairs the binding between ATAD2b and certain acetylated histone tails. Along with being found in humans, ATAD2b is also present in mice, bovine, zebra fish and rats. | ATAD2b (also known as KIAA1240) is an eukaryotic protein located in the nucleus that contains an AAA domain and a bromodomain. AAA ATPase domains use the energy of adenosine tri-phosphate (ATP) binding to participate in cellular activities as diverse as cell cycle control, signal transduction, disassembly of macromolecular complexes and regulation of gene expression. Bromodomains bind acetyl-lysine motifs and are thought to regulate protein–protein interactions in chromatin remodeling and transcriptional control. In humans, ATAD2B is an E2F target gene that binds to the MYC oncogene. High ATAD2b levels correlate with a higher risk of distant recurrence in breast cancer, and mutation of the bromodomain in ATAD2b impairs the binding between ATAD2b and certain acetylated histone tails. Along with being found in humans, ATAD2b is also present in mice, bovine, zebra fish and rats. | ||
| Line 9: | Line 9: | ||
Since much is unknown about ATAD2b we look to its paralogue ATAD2a which has been the focus of many studies. ATAD2a and ATAD2b both share the AAA and bromodomain, in which they are 97% and 74% identical respectively. identical and Based on the observed high sequence similarity with well characterized bromodomains, the ATAD2b bromodomain is expected to recognize acetylated lysine. However since no tests have yet to be done on this additional studies are needed to identify the exact modifications that the ATAD2b bromodomain may recognize. Throughout all bromodomains it seems as though there are very important residues that reside in the binding pocket, Tyr760, Tyr802 and Asn803. These three amino acids are highly conserved and thus must have a major impact in the binding pocket, they are most likely the key to bromodomains binding to acetylated lysine modifications. | Since much is unknown about ATAD2b we look to its paralogue ATAD2a which has been the focus of many studies. ATAD2a and ATAD2b both share the AAA and bromodomain, in which they are 97% and 74% identical respectively. identical and Based on the observed high sequence similarity with well characterized bromodomains, the ATAD2b bromodomain is expected to recognize acetylated lysine. However since no tests have yet to be done on this additional studies are needed to identify the exact modifications that the ATAD2b bromodomain may recognize. Throughout all bromodomains it seems as though there are very important residues that reside in the binding pocket, Tyr760, Tyr802 and Asn803. These three amino acids are highly conserved and thus must have a major impact in the binding pocket, they are most likely the key to bromodomains binding to acetylated lysine modifications. | ||
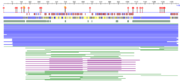
| - | [[Image:table 2.png |thumb| The entire ATAD2b protein sequenced using | + | [[Image:table 2.png |thumb|Figure 3. The entire ATAD2b protein sequenced using PredictProtein software. Diamonds represent protein binding, circles represent nucleosome binding. Red boxes are alpha helix, blue boxes beta sheet.]] |
== Sequence Analysis == | == Sequence Analysis == | ||
A BLAST search of the bromodomain in ATAD2b yields many other bromodomain-containing proteins. In the table below are 8 other proteins that contain a bromodomain. Their sequences were aligned to ATAD2b using Clustal Omega and show the percent identity and similarity to ATAD2b. You can see the conserved residues throughout all the proteins (marked with *), there are 13 out of the 71 residues or 18.3%. Many of the identical residues are a result of these 13 residues showing how highly conserved they are. They have very important function in the bromodomain which is why it would be expected to see the same residues in all bromodomains. The closest protein to ATAD2b is ATAD2a which is no surprise considering they are paralogues of each other. | A BLAST search of the bromodomain in ATAD2b yields many other bromodomain-containing proteins. In the table below are 8 other proteins that contain a bromodomain. Their sequences were aligned to ATAD2b using Clustal Omega and show the percent identity and similarity to ATAD2b. You can see the conserved residues throughout all the proteins (marked with *), there are 13 out of the 71 residues or 18.3%. Many of the identical residues are a result of these 13 residues showing how highly conserved they are. They have very important function in the bromodomain which is why it would be expected to see the same residues in all bromodomains. The closest protein to ATAD2b is ATAD2a which is no surprise considering they are paralogues of each other. | ||
| Line 44: | Line 44: | ||
8)Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52(9):747–55. | 8)Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52(9):747–55. | ||
| + | |||
| + | 9) Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; and others. PredictProtein---an open resource for online prediction of protein structural and functional features. Nucleic acids research, gku366. 2014. | ||
Revision as of 18:14, 11 December 2015
ATAD2b
| |||||||||||
References
1)Antonina Andreeva, Dave Howorth, Cyrus Chothia, Eugene Kulesha, Alexey Murzin, SCOP2 prototype: a new approach to protein structure mining (2014) Nucl. Acid Res., 42 (D1): D310-D314.
2)Cattaneo M., Morozumi Y., Perazza D. et al. (2014). Lessons from yeast on emerging roles of the ATAD2 protein family in gene regulation and genome organization. Mol. Cells 37, 851–856.
3)Chaikuad A., Petros A.M., Fedorov O. et al. (2014). Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain. Med. Chem. Commun. 5, 1843–1848.
4)Filippakopoulos P., Knapp S. (2014). Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 13, 337–356.
5)Filippakopoulos P., Picaud S., Mangos M. et al. (2012). Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231.
6)Wan W.N., Zhang Y.X., Wang X.M. et al. (2014). ATAD2 is highly expressed in ovarian carcinomas and indicates poor prognosis. Asian Pac. J. Cancer Prev. 15, 2777–2783.
7)Sanchez R., Meslamani J., Zhou M.M. (2014). The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta 1839, 676–685.
8)Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52(9):747–55.
9) Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; and others. PredictProtein---an open resource for online prediction of protein structural and functional features. Nucleic acids research, gku366. 2014.