We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Wabash13
From Proteopedia
(Difference between revisions)
| Line 36: | Line 36: | ||
<scene name='72/725338/Ribbon_diagram_n_c_rainbow/2'>Ribbon Diagram of Trypsin (N-->C Rainbow</scene> | <scene name='72/725338/Ribbon_diagram_n_c_rainbow/2'>Ribbon Diagram of Trypsin (N-->C Rainbow</scene> | ||
| + | |||
<scene name='Sandbox_45/Ctriadd102h57s195/4'>Catalytic Triad (Asp102, His57, Ser195)</scene> | <scene name='Sandbox_45/Ctriadd102h57s195/4'>Catalytic Triad (Asp102, His57, Ser195)</scene> | ||
| Line 41: | Line 42: | ||
---- | ---- | ||
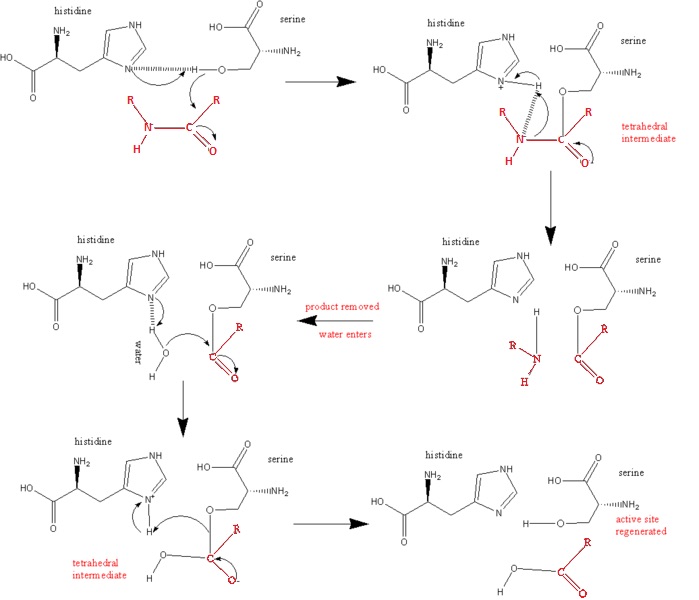
| - | + | '''Ser 195 nucleophilically attacks the scissile's peptide's carbonyl group''' | |
<scene name='72/725338/Serine__195/1'>Serine 195 - Base Catalysis Residue</scene> | <scene name='72/725338/Serine__195/1'>Serine 195 - Base Catalysis Residue</scene> | ||
| - | '''Ser 195 nucleophilically attacks the scissile's peptide's carbonyl group''' | ||
| - | <scene name='72/725338/His_57_asp_102/2'>Histidine 57 and Asp 102 </scene> | ||
'''The N3 of His 57 donates a proton (General Acid Catalysis) which is facilitated by the polarizing effect of Asp 102''' | '''The N3 of His 57 donates a proton (General Acid Catalysis) which is facilitated by the polarizing effect of Asp 102''' | ||
| + | <scene name='72/725338/His_57_asp_102/2'>Histidine 57 and Asp 102 </scene> | ||
| - | <scene name='72/725338/Aspartic_acid_102/2'>Aspartic Acid 102 - Important Residue in Stabilization of Catalytic Mechanism</scene> | ||
'''Asp 102 aids the process by its polarizing effect as an unsolved carboxylate ion which is hydrogen bonded to His 57''' | '''Asp 102 aids the process by its polarizing effect as an unsolved carboxylate ion which is hydrogen bonded to His 57''' | ||
| + | <scene name='72/725338/Aspartic_acid_102/2'>Aspartic Acid 102 - Important Residue in Stabilization of Catalytic Mechanism</scene> | ||
| + | |||
Revision as of 02:36, 19 February 2016
Trypsin Mechanism & Structure - Chase Francoeur, Elias Arellano
| |||||||||||