Factor IX
From Proteopedia
(Difference between revisions)
| Line 139: | Line 139: | ||
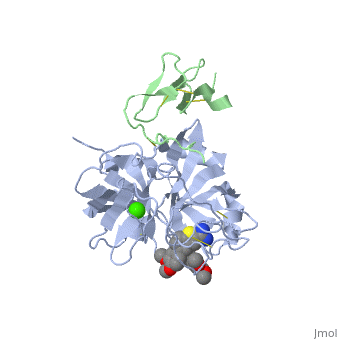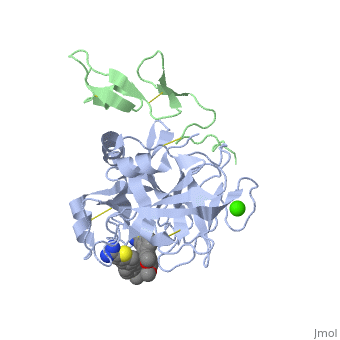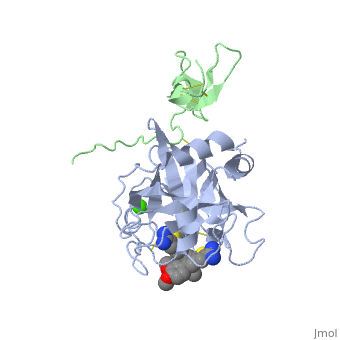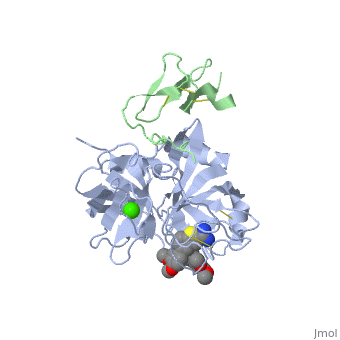
The Gla domain of Factor IX consists of an N-terminal loop and three short ''ω''-<scene name='Factor_IX/Antifactor_2/3'>helixes</scene>(helix A: residues 14-19; helix B: residues 24-32; helix C: residues 35-45). The calcium ions are positioned between the loop and the A and B helices with calcium liganding providing the folding energy to align the loop properly. Helices A and B are connected by a tight turn and further stabilized by a conserved disulfide bond between residues 18 and 23 of the Gla domain. The two cysteine residues also form two hydrogen bonds (Tyr-45 to Cys-18, 3.10 Å; Tyr-45 to Cys-23, 2.88 Å) to the side chain of a conserved Tyr-45 of helix C, providing an anchor to bundle three helices together. This tyrosine residue and the disulfide bond are part of the hydrophobic core between helix C and helix A/B that includes Phe-25, Ala-28, Phe-41, and Trp-42, a cluster that provides further hydrophobic energy to bind helix C to helix A/B. All of these residues are conserved among Gla domains of vitamin K-dependent proteins. | The Gla domain of Factor IX consists of an N-terminal loop and three short ''ω''-<scene name='Factor_IX/Antifactor_2/3'>helixes</scene>(helix A: residues 14-19; helix B: residues 24-32; helix C: residues 35-45). The calcium ions are positioned between the loop and the A and B helices with calcium liganding providing the folding energy to align the loop properly. Helices A and B are connected by a tight turn and further stabilized by a conserved disulfide bond between residues 18 and 23 of the Gla domain. The two cysteine residues also form two hydrogen bonds (Tyr-45 to Cys-18, 3.10 Å; Tyr-45 to Cys-23, 2.88 Å) to the side chain of a conserved Tyr-45 of helix C, providing an anchor to bundle three helices together. This tyrosine residue and the disulfide bond are part of the hydrophobic core between helix C and helix A/B that includes Phe-25, Ala-28, Phe-41, and Trp-42, a cluster that provides further hydrophobic energy to bind helix C to helix A/B. All of these residues are conserved among Gla domains of vitamin K-dependent proteins. | ||
| - | The binding of FIX to the phospholipid membrane is carried out by the loop, in the N terminus of the Gla domain. Four Ca2+ ions from Ca-2 to Ca-5 are essential to stabilize the ''ω''-loop region for membrane-binding <ref>PMID:8069632</ref> because these Ca2+ ions bind to conserved Gla7 and Gla8 residues in this region. The replacement by Mg2+ ions may lead to destabilization of the structure of the Gla domain, particularly in this region because of significant differences that exist with Ca2+ ion in both coordination distance and coordination sphere (Mg2+ ion takes generally a typical bipyramidal configuration and six coordination). This may explain why the Ca2+ ions are not replaced by Mg2+ ions in the present structure. | + | The binding of FIX to the phospholipid membrane is carried out by the loop, in the N terminus of the Gla domain. Four Ca2+ ions from Ca-2 to Ca-5 are essential to stabilize the ''ω''-loop region for membrane-binding <ref>PMID:8069632</ref> because these Ca2+ ions bind to conserved Gla7 and Gla8 residues in this region. The replacement by Mg2+ ions may lead to destabilization of the structure of the Gla domain, particularly in this region because of significant differences that exist with Ca2+ ion in both coordination distance and coordination sphere (Mg2+ ion takes generally a typical bipyramidal configuration and six coordination). This may explain why the Ca2+ ions are not replaced by Mg2+ ions in the present structure. See also [[Rotation of 4 degrees of FIX]]. |
The interaction of the Gla domain with the phospholipid membrane is carried out by binding to phosphatidylserine in the membrane through the interaction of calcium ions. Upon calcium binding to FIX a conformational change occurs in the Gla domain through the clustering of N-terminal hydrophobic residues into a hydrophobic patch, which is then exposed to the solvent. This hydrophobic patch then allows the association of the Gla domain with the cell surface membrane through electrostatic interactions between the phosphoserine head group and arginine and lysine residues in the Gla domain. The basic amino acid residues of factor IX (Lys-5 and Arg-10) bind to the glycerol phosphate backbone and the carboxyl group of the serine interacts with Ca-5 and Ca-6<ref>PMID:500729</ref> allowing membrane association. | The interaction of the Gla domain with the phospholipid membrane is carried out by binding to phosphatidylserine in the membrane through the interaction of calcium ions. Upon calcium binding to FIX a conformational change occurs in the Gla domain through the clustering of N-terminal hydrophobic residues into a hydrophobic patch, which is then exposed to the solvent. This hydrophobic patch then allows the association of the Gla domain with the cell surface membrane through electrostatic interactions between the phosphoserine head group and arginine and lysine residues in the Gla domain. The basic amino acid residues of factor IX (Lys-5 and Arg-10) bind to the glycerol phosphate backbone and the carboxyl group of the serine interacts with Ca-5 and Ca-6<ref>PMID:500729</ref> allowing membrane association. | ||
Revision as of 08:49, 21 February 2016
| |||||||||||
3D structures of factor IX
Updated on 21-February-2016
Additional Resources
For additional information, see: Colored & Bioluminescent Proteins
For additional information, see: Hemophilia
References
- ↑ Spitzer SG, Kuppuswamy MN, Saini R, Kasper CK, Birktoft JJ, Bajaj SP. Factor IXHollywood: substitution of Pro55 by Ala in the first epidermal growth factor-like domain. Blood. 1990 Oct 15;76(8):1530-7. PMID:2169923
- ↑ Schmidt AE, Bajaj SP. Structure-function relationships in factor IX and factor IXa. Trends Cardiovasc Med. 2003 Jan;13(1):39-45. PMID:12554099
- ↑ Zhong D, Bajaj MS, Schmidt AE, Bajaj SP. The N-terminal epidermal growth factor-like domain in factor IX and factor X represents an important recognition motif for binding to tissue factor. J Biol Chem. 2002 Feb 1;277(5):3622-31. Epub 2001 Nov 26. PMID:11723140 doi:10.1074/jbc.M111202200
- ↑ Perona JJ, Craik CS. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J Biol Chem. 1997 Nov 28;272(48):29987-90. PMID:9374470
- ↑ Ludwig M, Sabharwal AK, Brackmann HH, Olek K, Smith KJ, Birktoft JJ, Bajaj SP. Hemophilia B caused by five different nondeletion mutations in the protease domain of factor IX. Blood. 1992 Mar 1;79(5):1225-32. PMID:1346975
- ↑ Murav'ev IA, Mar'iasis ED, Starokozhko LE, Chebotarev VV, Krasova TG. [Determination of the biologic availability of preparations for topical use by experimental dermatologic methods] Farmatsiia. 1977 Jul-Aug;26(4):15-9. PMID:902786
- ↑ Morris DP, Stevens RD, Wright DJ, Stafford DW. Processive post-translational modification. Vitamin K-dependent carboxylation of a peptide substrate. J Biol Chem. 1995 Dec 22;270(51):30491-8. PMID:8530480
- ↑ Pan LC, Price PA. The propeptide of rat bone gamma-carboxyglutamic acid protein shares homology with other vitamin K-dependent protein precursors. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6109-13. PMID:3875856
- ↑ Knobloch JE, Suttie JW. Vitamin K-dependent carboxylase. Control of enzyme activity by the "propeptide" region of factor X. J Biol Chem. 1987 Nov 15;262(32):15334-7. PMID:2890628
- ↑ Knobloch JE, Suttie JW. Vitamin K-dependent carboxylase. Control of enzyme activity by the "propeptide" region of factor X. J Biol Chem. 1987 Nov 15;262(32):15334-7. PMID:2890628
- ↑ Cheung A, Engelke JA, Sanders C, Suttie JW. Vitamin K-dependent carboxylase: influence of the "propeptide" region on enzyme activity. Arch Biochem Biophys. 1989 Nov 1;274(2):574-81. PMID:2802629
- ↑ Lin PJ, Jin DY, Tie JK, Presnell SR, Straight DL, Stafford DW. The putative vitamin K-dependent gamma-glutamyl carboxylase internal propeptide appears to be the propeptide binding site. J Biol Chem. 2002 Aug 9;277(32):28584-91. Epub 2002 May 28. PMID:12034728 doi:10.1074/jbc.M202292200
- ↑ Sanford DG, Kanagy C, Sudmeier JL, Furie BC, Furie B, Bachovchin WW. Structure of the propeptide of prothrombin containing the gamma-carboxylation recognition site determined by two-dimensional NMR spectroscopy. Biochemistry. 1991 Oct 15;30(41):9835-41. PMID:1911775
- ↑ Lin PJ, Jin DY, Tie JK, Presnell SR, Straight DL, Stafford DW. The putative vitamin K-dependent gamma-glutamyl carboxylase internal propeptide appears to be the propeptide binding site. J Biol Chem. 2002 Aug 9;277(32):28584-91. Epub 2002 May 28. PMID:12034728 doi:10.1074/jbc.M202292200
- ↑ Furie B, Bouchard BA, Furie BC. Vitamin K-dependent biosynthesis of gamma-carboxyglutamic acid. Blood. 1999 Mar 15;93(6):1798-808. PMID:10068650
- ↑ Thompson AR. Structure, function, and molecular defects of factor IX. Blood. 1986 Mar;67(3):565-72. PMID:3511981
- ↑ Stenflo J, Fernlund P, Egan W, Roepstorff P. Vitamin K dependent modifications of glutamic acid residues in prothrombin. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2730-3. PMID:4528109
- ↑ Furie B, Furie BC. Molecular and cellular biology of blood coagulation. N Engl J Med. 1992 Mar 19;326(12):800-6. PMID:1538724 doi:http://dx.doi.org/10.1056/NEJM199203193261205
- ↑ Shikamoto Y, Morita T, Fujimoto Z, Mizuno H. Crystal structure of Mg2+- and Ca2+-bound Gla domain of factor IX complexed with binding protein. J Biol Chem. 2003 Jun 27;278(26):24090-4. Epub 2003 Apr 14. PMID:12695512 doi:10.1074/jbc.M300650200
- ↑ Huang M, Furie BC, Furie B. Crystal structure of the calcium-stabilized human factor IX Gla domain bound to a conformation-specific anti-factor IX antibody. J Biol Chem. 2004 Apr 2;279(14):14338-46. Epub 2004 Jan 13. PMID:14722079 doi:10.1074/jbc.M314011200
- ↑ Golinelli-Pimpaneau B, Gigant B, Bizebard T, Navaza J, Saludjian P, Zemel R, Tawfik DS, Eshhar Z, Green BS, Knossow M. Crystal structure of a catalytic antibody Fab with esterase-like activity. Structure. 1994 Mar 15;2(3):175-83. PMID:8069632
- ↑ Furie BC, Blumenstein M, Furie B. Metal binding sites of a gamma-carboxyglutamic acid-rich fragment of bovine prothrombin. J Biol Chem. 1979 Dec 25;254(24):12521-30. PMID:500729
- ↑ Nakagawa T, Tani M, Kita K, Ito M. Preparation of fluorescence-labeled GM1 and sphingomyelin by the reverse hydrolysis reaction of sphingolipid ceramide N-deacylase as substrates for assay of sphingolipid-degrading enzymes and for detection of sphingolipid-binding proteins. J Biochem. 1999 Sep;126(3):604-11. PMID:10467178
Proteopedia Page Contributors and Editors (what is this?)
Nadia Dorochko, Michal Harel, Alexander Berchansky, David Canner