This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1178
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
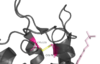
[[Image:Disulfide_Bond_Ray_Traced.png|100 px|left|thumb|Figure Legend]] | [[Image:Disulfide_Bond_Ray_Traced.png|100 px|left|thumb|Figure Legend]] | ||
= TAK-875 Binding = | = TAK-875 Binding = | ||
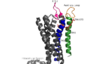
| - | Tak-875 is known to be a [https://en.wikipedia.org/wiki/Partial_agonist partial agonist] of GPR40. The bonding of this ligand to the bonding site is fairly unique, as it is proposed that the ligand must enter through the [https://en.wikipedia.org/wiki/Cell_membrane membrane bilayer]. This is performed via a method similar to ligand binding to sphingosine 1-phosphate receptor 1 [[:3v2w]], retinal loading of opsin [[:4j4q]] and the entry of anandamide in [https://en.wikipedia.org/wiki/Cannabinoid_receptor cannabinoid receptors] | + | Tak-875 is known to be a [https://en.wikipedia.org/wiki/Partial_agonist partial agonist] of GPR40. The bonding of this ligand to the bonding site is fairly unique, as it is proposed that the ligand must enter through the [https://en.wikipedia.org/wiki/Cell_membrane membrane bilayer]. This is performed via a method similar to ligand binding to sphingosine 1-phosphate receptor 1 [[:3v2w]], retinal loading of opsin [[:4j4q]] and the entry of anandamide in [https://en.wikipedia.org/wiki/Cannabinoid_receptor cannabinoid receptors], in which extracellular loops block the binding from the extracellular matrix (Hanson Et al., 2012). In contrast, delta opioid receptor binding [[:4ej4]] allow for binding directly from the [https://en.wikipedia.org/wiki/Extracellular_matrix extracellular matrix]. The binding mechanism through the bilayer may be selectively favoring the free fatty acid because of the [https://en.wikipedia.org/wiki/Chemical_polarity#Nonpolar_molecules non-polar] regions of the ligand. |
== Disease == | == Disease == | ||
Current revision
GPR40 bound to TAK-875
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644