Lac repressor
From Proteopedia
(Difference between revisions)
| Line 96: | Line 96: | ||
== 3D structures of Lac repressor== | == 3D structures of Lac repressor== | ||
| - | [[Lac repressor | + | *Lac repressor |
| + | |||
| + | **[[3edc]] – EcLAC + hexanediol - ''Escherichia coli''<br /> | ||
| + | **[[2pe5]] – EcLAC residues 2-331 (mutant) + effector<br /> | ||
| + | **[[1lbh]] - EcLAC + effector<br /> | ||
| + | **[[2p9h]] - EcLAC residues 62-330 + effector<br /> | ||
| + | **[[2paf]] - EcLAC residues 62-330 + anti-inducer<br /> | ||
| + | **[[1lbi]] – EcLAC <br /> | ||
| + | **[[1jye]], [[1jyf]] - EcLAC (mutant)<br /> | ||
| + | **[[1lqc]] - EcLAC headpiece – NMR<br /> | ||
| + | **[[1tlf]] - EcLAC residues 19-319<br /> | ||
| + | **[[2r2v]] – LAC coiled-coil - yeast | ||
| + | |||
| + | *Lac repressor complex with DNA | ||
| + | |||
| + | **[[2kei]], [[1l1m]] – EcLAC DNA-binding domain (mutant) + O1 operator –NMR<BR /> | ||
| + | **[[2kej]] - EcLAC DNA-binding domain (mutant) + O2 operator – NMR<BR /> | ||
| + | **[[2kek]] - EcLAC DNA-binding domain (mutant) + O3 operator – NMR<BR /> | ||
| + | **[[2bjc]] - EcLAC DNA-binding domain (mutant) + GAL operator – NMR<BR /> | ||
| + | **[[1osl]] - EcLAC DNA-binding domain (mutant) + DNA – NMR<BR /> | ||
| + | **[[1cjg]], [[1lcc]], [[1lcd]] - EcLAC headpiece + DNA – NMR<br /> | ||
| + | **[[1jwl]] - EcLAC + O1 operator + effector<br /> | ||
| + | **[[1lbg]] - EcLAC + DNA + inducer<br /> | ||
| + | **[[1efa]] - EcLAC residues 1-333 (mutant) + DNA | ||
| + | }} | ||
==See Also== | ==See Also== | ||
Revision as of 07:53, 11 April 2016
Contents |
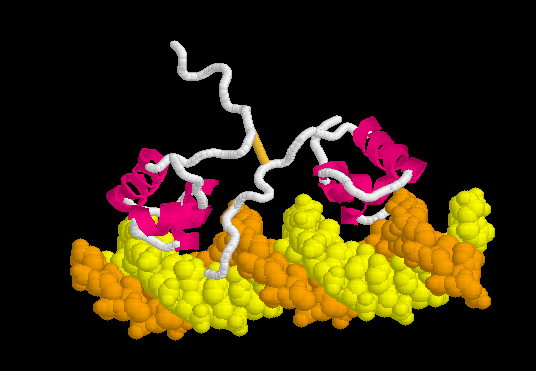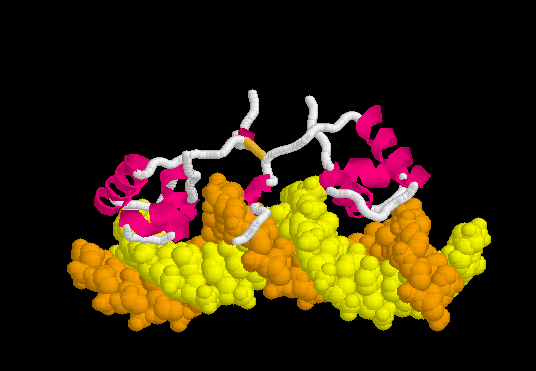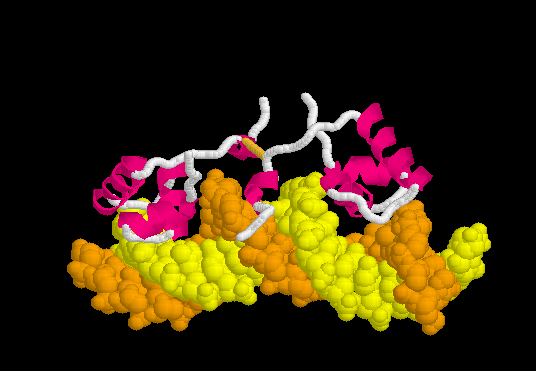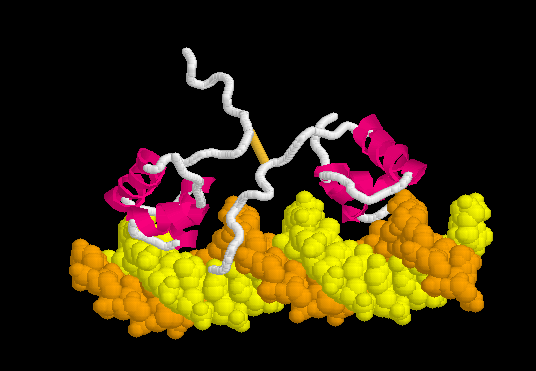
Lac Repressor
| |||||||||||
3D structures of Lac repressor
- Lac repressor
- 3edc – EcLAC + hexanediol - Escherichia coli
- 2pe5 – EcLAC residues 2-331 (mutant) + effector
- 1lbh - EcLAC + effector
- 2p9h - EcLAC residues 62-330 + effector
- 2paf - EcLAC residues 62-330 + anti-inducer
- 1lbi – EcLAC
- 1jye, 1jyf - EcLAC (mutant)
- 1lqc - EcLAC headpiece – NMR
- 1tlf - EcLAC residues 19-319
- 2r2v – LAC coiled-coil - yeast
- 3edc – EcLAC + hexanediol - Escherichia coli
- Lac repressor complex with DNA
- 2kei, 1l1m – EcLAC DNA-binding domain (mutant) + O1 operator –NMR
- 2kej - EcLAC DNA-binding domain (mutant) + O2 operator – NMR
- 2kek - EcLAC DNA-binding domain (mutant) + O3 operator – NMR
- 2bjc - EcLAC DNA-binding domain (mutant) + GAL operator – NMR
- 1osl - EcLAC DNA-binding domain (mutant) + DNA – NMR
- 1cjg, 1lcc, 1lcd - EcLAC headpiece + DNA – NMR
- 1jwl - EcLAC + O1 operator + effector
- 1lbg - EcLAC + DNA + inducer
- 1efa - EcLAC residues 1-333 (mutant) + DNA
- 2kei, 1l1m – EcLAC DNA-binding domain (mutant) + O1 operator –NMR
}}
See Also
- DNA-protein interactions, an overview introducing helix-turn-helix, leucine zipper, and zinc finger proteins.
- Category: Lac repressor and Category: Lac Repressor, automatically-generated pages that list PDB codes for lac repressor models.
- Morphs where the morph of the lac repressor is used as an example.
- Lac repressor morph methods
- See: Regulation of Gene Expression for additional mechanisms of Gene Regulation
- For additional information, see: Transcription and RNA Processing
References & Notes
- ↑ L'opéron: groupe de gènes à expression coordonée par un opérateur. [Operon: a group of genes with the expression coordinated by an operator.] C R Hebd Seances Acad Sci., 250:1727-9, 1960. PubMed 14406329
- ↑ The lac repressor. Lewis, M. C R Biol. 328:521-48, 2005. PubMed 15950160
- ↑ This domain coloring scheme is adapted from Fig. 6 in the review by Lewis (C. R. Biol. 328:521, 2005). Domains are 1-45, 46-62, (63-162,291-320), (163-290,321-332), 330-339, and 340-357.
- ↑ Conservation results for 1lbg are from the precalculated ConSurf Database, using 103 sequences from Swiss-Prot with an average pairwise distance of 2.4.
- ↑ Conservation results for 1lbi are from the ConSurf Server, using 100 sequences from Uniprot with an average pairwise distance of 1.3.
- ↑ 6.0 6.1 For these scenes, the 20-model PDB files for 1osl and 1l1m were reduced in size, to avoid exceeding the java memory available to the Jmol applet. All atoms except amino acid alpha carbons and DNA phosphorus atoms were removed using the free program alphac.exe from PDBTools. Secondary structure HELIX records from the original PDB file header were retained. The results are Image:1osl ca.pdb and Image:1l1m ca.pdb.
- ↑ Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012 Jun 22;336(6088):1595-8. PMID:22723426 doi:10.1126/science.1221648
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233-69. PMID:20334529 doi:10.1146/annurev-biochem-060408-091030
- ↑ Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007 Nov 2;131(3):530-43. PMID:17981120 doi:10.1016/j.cell.2007.09.024
- ↑ 10.0 10.1 10.2 Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009 Oct 29;461(7268):1248-53. PMID:19865164 doi:10.1038/nature08473
- ↑ Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011 Feb 24;470(7335):498-502. Epub 2011 Jan 26. PMID:21270796 doi:10.1038/nature09775
- ↑ Honig B, Rohs R. Biophysics: Flipping Watson and Crick. Nature. 2011 Feb 24;470(7335):472-3. PMID:21350476 doi:10.1038/470472a
- ↑ Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010 Apr;17(4):423-9. Epub 2010 Apr 4. PMID:20364130 doi:10.1038/nsmb.1800
- ↑ Powerpoint is a registered trademark for a software package licensed by Microsoft Corp..
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Michal Harel, Alexander Berchansky, Joel L. Sussman, Karsten Theis, Henry Jakubowski, David Canner, Eran Hodis