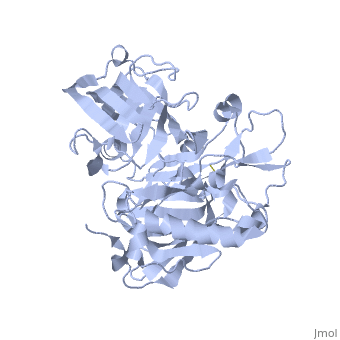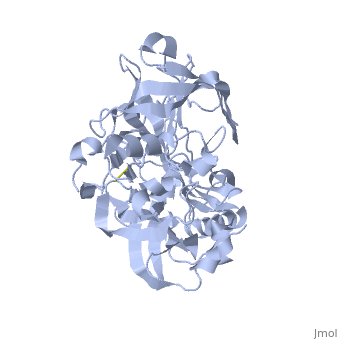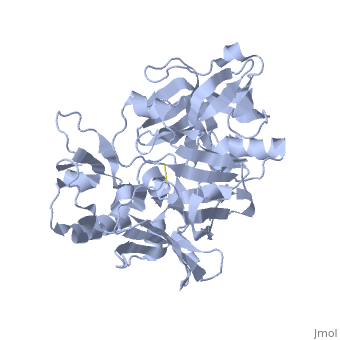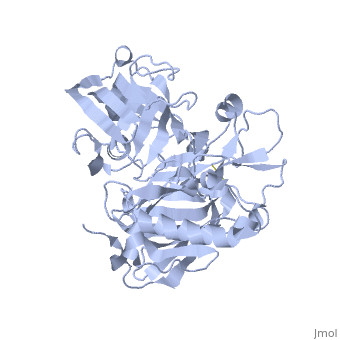Beta Secretase (BACE1) 1SGZ
From Proteopedia
| Line 1: | Line 1: | ||
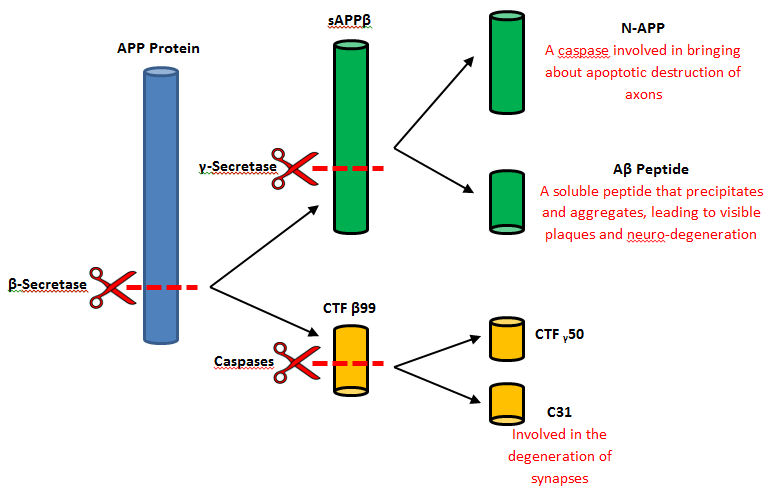
| - | Beta-secretase, also known as '''BACE''' or '''Memapsin 2''', is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing [[amyloid precursor protein]] (APP), which is an integral membrane protein. A malfunction in the processing of APP results in the formation of the peptide [[amyloid beta]]. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with [[Alzheimers Disease]]. '''Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.''' | + | Beta-secretase, also known as '''BACE''' or '''Memapsin 2''', is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing [[amyloid precursor protein]] (APP), which is an integral membrane protein.<ref>1. Ghosh, A.; Kumaragurubaran, N.; Hong, L.; Koelsh, G.; Tang, J. Memapsin 2 (Beta-Secretase) Inhibitors: Drug Development. CAR Current Alzheimer Research. [Online] 2008, 5, 121–131. |
| + | http://jamesmadisonva.library.ingentaconnect.com/content/ben/car/2008/00000005/00000002/art00004</ref> A malfunction in the processing of APP results in the formation of the peptide [[amyloid beta]]. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with [[Alzheimers Disease]]. '''Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.''' | ||
| Line 7: | Line 8: | ||
== Structure == | == Structure == | ||
<StructureSection load='1SGZ' size='340' side='right' caption='Human Beta-secretase (PDB 1SGZ)' scene=''> | <StructureSection load='1SGZ' size='340' side='right' caption='Human Beta-secretase (PDB 1SGZ)' scene=''> | ||
| - | BACE-1 is a type-1 integral membrane glycoprotein that is affected by the positions on the outer domains of amino acids. Cleavage of soluble substrates is possible in external environment due to the enzyme’s high values of Km failing to observe in the internal environment. BACE-1 prefers the cleavage of longer substrates over smaller peptide based substrates due to approximate acidic pH of 4.5 resulting from the cleavage. The acidic and polar environment of the enzyme allow a wide variety of substrates to bind to the membrane. The two catalytic aspartate residues that reside in the active site of BACE-1 are Asp 32 and Asp 228. The active site of BACE-1 has a flexible antiparallel -hairpin, also known as a flap, which is found in the residues of 67 to 77, that controls the accessibility of the substrate to the active site, and the flap also has to set the substrate in the correct orientation for catalysis. In an open conformation, the active site allows the substrate to enter without any difficulty and regulates its activity. In an inactive form, the binding affinity to the inhibitor is decreased, which then lowers the substrate binding and enzymatic activity. For the closed conformation, the flap with the inhibitor bound is firmly secured in place. | ||
| - | + | BACE-1 is a type-1 integral membrane glycoprotein that is affected by the positions on the outer domains of amino acids (7) . BACE-1 prefers the cleavage of longer substrates over smaller peptide based substrates due to approximate acidic pH of 4.5 resulting from the cleavage. The acidic and polar environment of the enzyme allow a wide variety of substrates to bind to the membrane. Cleavage of soluble substrates is possible in external environment due to the enzyme’s high values of Km failing to observe in the internal environment (8). The two catalytic aspartate residues that reside in the active site of BACE-1 are Asp 32 and Asp 228 (7). The active site of BACE-1 has a flexible antiparallel -hairpin, also known as a flap, which is found in the residues of 67 to 77, that controls the accessibility of the substrate to the active site, and the flap also has to set the substrate in the correct orientation for catalysis (8). In an open conformation, the active site allows the substrate to enter without any difficulty and regulates its activity. In an inactive form, the binding affinity to the inhibitor is decreased, which then lowers the substrate binding and enzymatic activity. For the closed conformation, the flap with the inhibitor bound is firmly secured in place (8). | |
| - | + | The S3 pocket is adjacent to the Thr 232 and the amino acids from the 10s loop, which contains Glycine and Glutamine. The Glycine located in the 10s loop that the substrate forms a hydrogen bond with helps stabilize the 10s loop which further balances the interaction of the beta secretase-substrate. In an open conformation of the 10s loop, the substrate and the S3 pocket have a greater affinity to bind with each other (9). The stabilization of the structure depends on the active site, flap and 10s loop. | |
| + | Specifically, BACE-1 is a transmembrane aspartic protease which cleaves the amyloid-beta precursor protein and produces the amyloid-beta peptide. Two water molecules are in the process for the aspartic protease. The location of the first water molecule for BACE-1 is between the Asp 32 and Asp 228 (8).Once the substrate binds, the two aspartate residues form a hydrogen bond with the water molecule. Then, the water molecule becomes the nucleophile that attacks the carbonyl. The second water molecule forms a hydrogen bond with Tyr residue that is found in the flap. The behavior of the two water molecules affects the activity of BACE-1 (8). | ||
== Structural Highlights == | == Structural Highlights == | ||
Revision as of 02:00, 13 April 2016
Beta-secretase, also known as BACE or Memapsin 2, is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing amyloid precursor protein (APP), which is an integral membrane protein.[1] A malfunction in the processing of APP results in the formation of the peptide amyloid beta. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with Alzheimers Disease. Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.
Enzyme Class
Beta-secretase is an enzyme that is classified as a class 3 enzyme, which are hydrolases. The enzyme acts on breaking peptide bonds and therefore is also considered a peptidase and belongs to the subclass of aspartic acid endopeptidases.
Structure
| |||||||||||
References
- ↑ 1. Ghosh, A.; Kumaragurubaran, N.; Hong, L.; Koelsh, G.; Tang, J. Memapsin 2 (Beta-Secretase) Inhibitors: Drug Development. CAR Current Alzheimer Research. [Online] 2008, 5, 121–131. http://jamesmadisonva.library.ingentaconnect.com/content/ben/car/2008/00000005/00000002/art00004