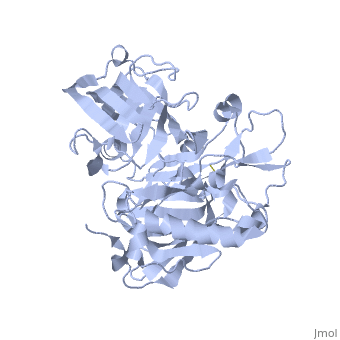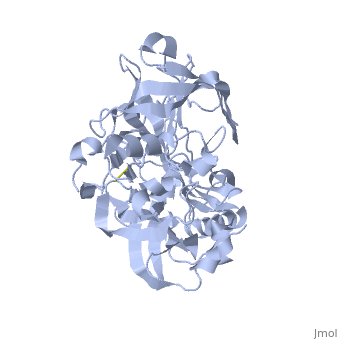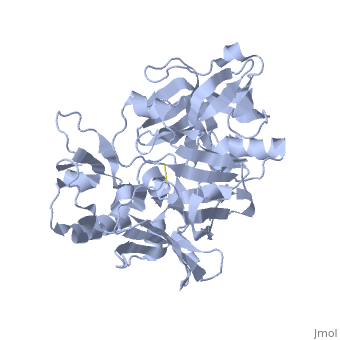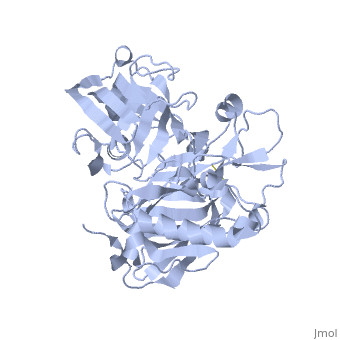Beta Secretase (BACE1) 1SGZ
From Proteopedia
| Line 26: | Line 26: | ||
http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831</ref>. | http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831</ref>. | ||
| - | Specifically, BACE-1 is a transmembrane aspartic protease which cleaves the amyloid-beta precursor protein and produces the amyloid-beta peptide. Two water molecules are in the process for the aspartic protease. The location of the first water molecule for BACE-1 is between the Asp 32 and Asp 228 | + | Specifically, BACE-1 is a transmembrane aspartic protease which cleaves the amyloid-beta precursor protein and produces the amyloid-beta peptide. Two water molecules are in the process for the aspartic protease. The location of the first water molecule for BACE-1 is between the Asp 32 and Asp 228. Once the substrate binds, the two aspartate residues form a hydrogen bond with the water molecule. Then, the water molecule becomes the nucleophile that attacks the carbonyl. The second water molecule forms a hydrogen bond with Tyr residue that is found in the flap. The behavior of the two water molecules affects the activity of BACE-1 <ref name="seven">Shimizu, H.; Tosaki, A.; Kaneko, K.; Hisano, T.; Sakurai, T.; Nukina, N. Crystal Structure of an Active Form of BACE1, an Enzyme Responsible for Amyloid Beta Protein Production. Molecular and Cellular Biology [Online] 2008, 28(11), 3663-671. |
http://mcb.asm.org/content/28/11/3663.full.pdf+html | http://mcb.asm.org/content/28/11/3663.full.pdf+html | ||
</ref>. | </ref>. | ||
| Line 35: | Line 35: | ||
The mechanism of this protease activity is an acid-base reaction, common in organic chemistry. The two aspartate molecules coordinate a water molecule through hydrogen bonding. The aspartate molecule takes a hydrogen from a water molecule, which activates the water as a good nucleophile. The good water nucleophile attacks the carbonyl carbon in the peptide bond with the attack forms a tetrahedral intermediate. A pair of electrons is moved to re-create a carbonyl group which breaks the peptide bond and re-generates an aspartic oxyanion. No covalent bonds are formed between the beta-secretase side chains and the substrate on the enzyme and therefore the fragments can exit the active site easily. | The mechanism of this protease activity is an acid-base reaction, common in organic chemistry. The two aspartate molecules coordinate a water molecule through hydrogen bonding. The aspartate molecule takes a hydrogen from a water molecule, which activates the water as a good nucleophile. The good water nucleophile attacks the carbonyl carbon in the peptide bond with the attack forms a tetrahedral intermediate. A pair of electrons is moved to re-create a carbonyl group which breaks the peptide bond and re-generates an aspartic oxyanion. No covalent bonds are formed between the beta-secretase side chains and the substrate on the enzyme and therefore the fragments can exit the active site easily. | ||
| - | [[Image:BsecretaseActiveSiteMechanism.png]] | + | [[Image:BsecretaseActiveSiteMechanism.png]] <ref>Sugana, K., Padlan E., Smith, C., Carlson, W., and Davis, D. (1987). Retrieved April 5, 2016 from https://upload.wikimedia.org/wikipedia/commons/thumb/0/00/Aspartyl_protease_mechanism.png/510px-Aspartyl_protease_mechanism.png</ref> |
== Function == | == Function == | ||
Revision as of 02:24, 13 April 2016
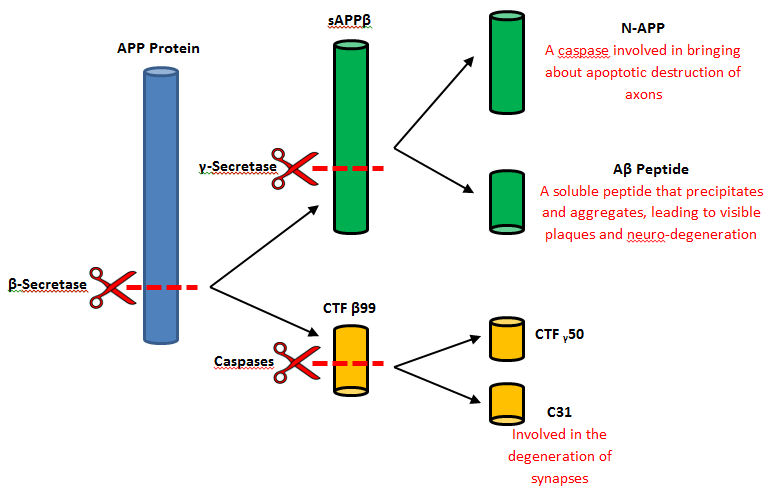
Beta-secretase, also known as BACE or Memapsin 2, is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing amyloid precursor protein (APP), which is an integral membrane protein.[1] A malfunction in the processing of APP results in the formation of the peptide amyloid beta. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with Alzheimers Disease. [2] Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.[3]
Enzyme Class
Beta-secretase is an enzyme that is classified as a class 3 enzyme, which are hydrolases [4]. The enzyme acts on breaking peptide bonds and therefore is also considered a peptidase and belongs to the subclass of aspartic acid endopeptidases.[5]
Structure
| |||||||||||
References
- ↑ Ghosh, A.; Kumaragurubaran, N.; Hong, L.; Koelsh, G.; Tang, J. Memapsin 2 (Beta-Secretase) Inhibitors: Drug Development. CAR Current Alzheimer Research. [Online] 2008, 5, 121–131. http://jamesmadisonva.library.ingentaconnect.com/content/ben/car/2008/00000005/00000002/art00004
- ↑ John, V.; Beck, J. P.; Bienkowski, M. J.; Sinha, S.; Heinrikson, R. L. Human β-Secretase (BACE) And BACE Inhibitors. ChemInform. [Online] 2004, 35, 4625-4630. http://pubs.acs.org/doi/pdf/10.1021/jm030247h
- ↑ Hu, X.; Hicks, C. W.; He, W.; Wong, P.; Macklin, W. B.; Trapp, B. D.; Yan, R. Bace1 Modulates Myelination in the Central and Peripheral Nervous System. Nature Neuroscience Nat Neurosci. [Online] 2006, 9, 1520–1525. http://www.nature.com/neuro/journal/v9/n12/abs/nn1797.html
- ↑ Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. The Enzyme List Class 3 — Hydrolases. http://www.enzyme-database.org/downloads/ec3.pdf
- ↑ DBGET Search. KEGG ENZYME: 3.4.23.46. http://www.genome.jp/dbget-bin/www_bget?ec:3.4.23.46
- ↑ 6.0 6.1 Venugopal, C.; Demos, C. M.; Rao, K. S. J.; Pappolla, M. A.; Sambamurti, K. Beta-secretase: Structure, Function, and Evolution. CNS & Neurological Disorders Drug Targets. [Online] 2008. 7(3), 278–294. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2921875/
- ↑ 7.0 7.1 7.2 Shimizu, H.; Tosaki, A.; Kaneko, K.; Hisano, T.; Sakurai, T.; Nukina, N. Crystal Structure of an Active Form of BACE1, an Enzyme Responsible for Amyloid Beta Protein Production. Molecular and Cellular Biology [Online] 2008, 28(11), 3663-671. http://mcb.asm.org/content/28/11/3663.full.pdf+html
- ↑ Kumalo, M.; Soumendranath B.; Soliman, M. E. Investigation of Flap Flexibility of β-secretase Using Molecular Dynamic Simulations. Journal of Biomolecular Structure and Dynamics [Online] , 2015, 1-12. http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831
- ↑ Sugana, K., Padlan E., Smith, C., Carlson, W., and Davis, D. (1987). Retrieved April 5, 2016 from