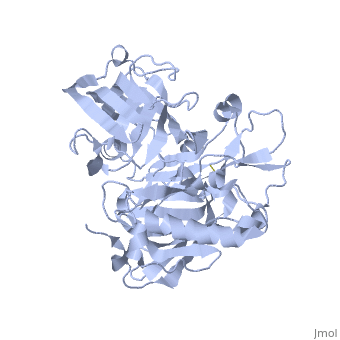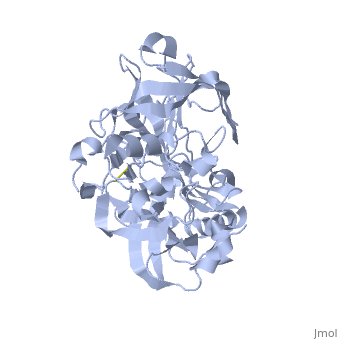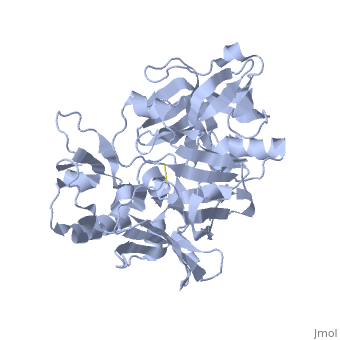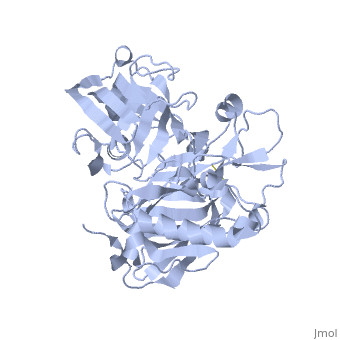Beta Secretase (BACE1) 1SGZ
From Proteopedia
| Line 52: | Line 52: | ||
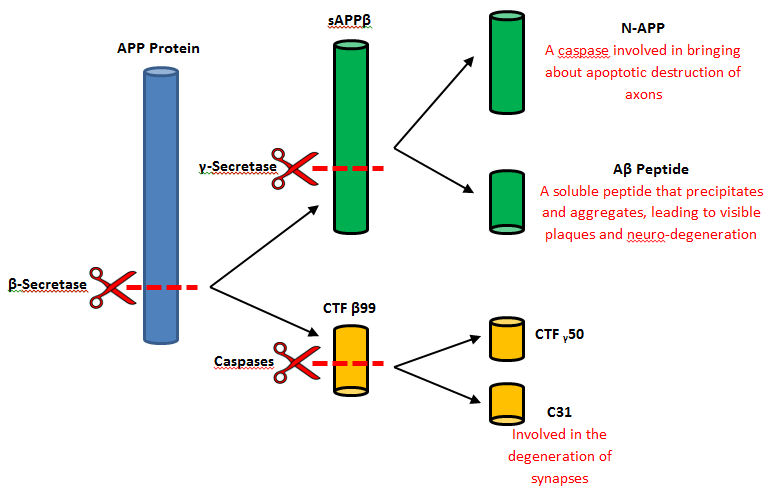
AD occurs because of plaques which are formed from a buildup of amyloid beta. Amyloid beta (AB) is a four kilodalton protein made up of about 39-43 amino acids in length, which aggregates in the brain and causes the destruction of the brain <ref name="fourteen>Shoji, M.; Golde, T.; Ghiso, J.; Cheung, T.; Estus, S.; Shaffer, L.; Cai, X.; Mckay, D.; Tintner, R.; Frangione, B.; Et, A. Production Of the Alzheimer Amyloid Beta Protein by Normal Proteolytic Processing. Science. 1992, 258, 126–129. | AD occurs because of plaques which are formed from a buildup of amyloid beta. Amyloid beta (AB) is a four kilodalton protein made up of about 39-43 amino acids in length, which aggregates in the brain and causes the destruction of the brain <ref name="fourteen>Shoji, M.; Golde, T.; Ghiso, J.; Cheung, T.; Estus, S.; Shaffer, L.; Cai, X.; Mckay, D.; Tintner, R.; Frangione, B.; Et, A. Production Of the Alzheimer Amyloid Beta Protein by Normal Proteolytic Processing. Science. 1992, 258, 126–129. | ||
| - | </ref>. Amyloid beta is cleaved from the membrane protein APP done so by two proteases, beta-secretase and gamma-secretase. Amyloid betas are amphiphilic peptides with a hydrophilic (water loving) N-terminal side (residues 1-28) and a hydrophobic (water hating) C-terminal side (residues 29-40). These characteristics of the AB is what affects the toxicity of the peptide aggregates in the brain. | + | </ref>. Amyloid beta is cleaved from the membrane protein APP done so by two proteases, beta-secretase and gamma-secretase. Amyloid betas are amphiphilic peptides with a hydrophilic (water loving) N-terminal side (residues 1-28) and a hydrophobic (water hating) C-terminal side (residues 29-40). These characteristics of the AB is what affects the toxicity of the peptide aggregates in the brain<ref name="fifteen"></ref>. |
== Inhibitors == | == Inhibitors == | ||
| - | In recent years there has been much work done with beta-secretase inhibitors and synthesizing therapeutic drugs that will inhibit the production of beta-amyloids. An area that has specifically been studied is Beta-secretase processing of APP. Processing of APP is the first step in the synthesis of amyloid-beta, and therefore inhibiting it could be a successful way to inhibit the development of Alzheimer's diseases. Two main types of beta-secretase inhibitors that are being studied include; peptidomimetic beta-secretase inhibitors and nonpeptidomimetic beta-secretase inhibitors. | + | In recent years there has been much work done with beta-secretase inhibitors and synthesizing therapeutic drugs that will inhibit the production of beta-amyloids. An area that has specifically been studied is Beta-secretase processing of APP. Processing of APP is the first step in the synthesis of amyloid-beta, and therefore inhibiting it could be a successful way to inhibit the development of Alzheimer's diseases<ref name="sixteen">Thompson, L.; Bronson, J.; Zusi, C.; Progress in the discovery of BACE Inhibitors. Current Pharmaceutical design. 2005, 11, 3383-3404. |
| + | </ref>. Two main types of beta-secretase inhibitors that are being studied include; peptidomimetic beta-secretase inhibitors and nonpeptidomimetic beta-secretase inhibitors. | ||
Peptidomimetic beta-secretase inhibitors focus on substrate-based inhibition and were created by analyzing the specificity and kinetics of the beta-secretase enzyme. One specific development of a peptidomimetic beta-secretase inhibitor, by the Elan/Pharmacia team, involves a cell-permeable, dose-dependent, mechanism-specific reduction of the harmful amyloid-beta substance in human embryonic cells. Identification of a specific sequence, P16 to P5’, in beta-secretase substrate was the key discovery that led to the development of a peptidomimetic beta-secretase inhibitor by replacing the P1 residue with a statine, which in uncleavable, and replacing the P1’ Asp with valine. The new analogue created was then used to create purified beta-secretase that was sequenced and cloned. The N-terminus and the C-terminus of the created and purified beta-secretase were truncated to create a small peptide inhibitor, which was then transformed into a cell-permeable peptidomimetic beta-secretase inhibitor by dividing it into three regions. The regions included; an N-terminus, a C-terminus, and central core section that included statine. The Elan targeted these three regions and focused on modifying them to have less peptidic qualities but still keep beta-secretase enzyme activity<ref name="two">John, V.; Beck, J. P.; Bienkowski, M. J.; Sinha, S.; Heinrikson, R. L. Human β-Secretase (BACE) And BACE Inhibitors. ChemInform. [Online] 2004, 35, 4625-4630. | Peptidomimetic beta-secretase inhibitors focus on substrate-based inhibition and were created by analyzing the specificity and kinetics of the beta-secretase enzyme. One specific development of a peptidomimetic beta-secretase inhibitor, by the Elan/Pharmacia team, involves a cell-permeable, dose-dependent, mechanism-specific reduction of the harmful amyloid-beta substance in human embryonic cells. Identification of a specific sequence, P16 to P5’, in beta-secretase substrate was the key discovery that led to the development of a peptidomimetic beta-secretase inhibitor by replacing the P1 residue with a statine, which in uncleavable, and replacing the P1’ Asp with valine. The new analogue created was then used to create purified beta-secretase that was sequenced and cloned. The N-terminus and the C-terminus of the created and purified beta-secretase were truncated to create a small peptide inhibitor, which was then transformed into a cell-permeable peptidomimetic beta-secretase inhibitor by dividing it into three regions. The regions included; an N-terminus, a C-terminus, and central core section that included statine. The Elan targeted these three regions and focused on modifying them to have less peptidic qualities but still keep beta-secretase enzyme activity<ref name="two">John, V.; Beck, J. P.; Bienkowski, M. J.; Sinha, S.; Heinrikson, R. L. Human β-Secretase (BACE) And BACE Inhibitors. ChemInform. [Online] 2004, 35, 4625-4630. | ||
Revision as of 03:10, 13 April 2016
Beta-secretase, also known as BACE or Memapsin 2, is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing amyloid precursor protein (APP), which is an integral membrane protein.[1] A malfunction in the processing of APP results in the formation of the peptide amyloid beta. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with Alzheimers Disease. [2] Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.[3]
Enzyme Class
Beta-secretase is an enzyme that is classified as a class 3 enzyme, which are hydrolases [4]. The enzyme acts on breaking peptide bonds and therefore is also considered a peptidase and belongs to the subclass of aspartic acid endopeptidases.[5]
Structure
| |||||||||||
References
- ↑ Ghosh, A.; Kumaragurubaran, N.; Hong, L.; Koelsh, G.; Tang, J. Memapsin 2 (Beta-Secretase) Inhibitors: Drug Development. CAR Current Alzheimer Research. [Online] 2008, 5, 121–131. http://jamesmadisonva.library.ingentaconnect.com/content/ben/car/2008/00000005/00000002/art00004
- ↑ 2.0 2.1 2.2 John, V.; Beck, J. P.; Bienkowski, M. J.; Sinha, S.; Heinrikson, R. L. Human β-Secretase (BACE) And BACE Inhibitors. ChemInform. [Online] 2004, 35, 4625-4630. http://pubs.acs.org/doi/pdf/10.1021/jm030247h
- ↑ Hu, X.; Hicks, C. W.; He, W.; Wong, P.; Macklin, W. B.; Trapp, B. D.; Yan, R. Bace1 Modulates Myelination in the Central and Peripheral Nervous System. Nature Neuroscience Nat Neurosci. [Online] 2006, 9, 1520–1525. http://www.nature.com/neuro/journal/v9/n12/abs/nn1797.html
- ↑ Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. The Enzyme List Class 3 — Hydrolases. http://www.enzyme-database.org/downloads/ec3.pdf
- ↑ DBGET Search. KEGG ENZYME: 3.4.23.46. http://www.genome.jp/dbget-bin/www_bget?ec:3.4.23.46
- ↑ 6.0 6.1 6.2 Venugopal, C.; Demos, C. M.; Rao, K. S. J.; Pappolla, M. A.; Sambamurti, K. Beta-secretase: Structure, Function, and Evolution. CNS & Neurological Disorders Drug Targets. [Online] 2008. 7(3), 278–294. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2921875/
- ↑ 7.0 7.1 7.2 Shimizu, H.; Tosaki, A.; Kaneko, K.; Hisano, T.; Sakurai, T.; Nukina, N. Crystal Structure of an Active Form of BACE1, an Enzyme Responsible for Amyloid Beta Protein Production. Molecular and Cellular Biology [Online] 2008, 28(11), 3663-671. http://mcb.asm.org/content/28/11/3663.full.pdf+html
- ↑ Kumalo, M.; Soumendranath B.; Soliman, M. E. Investigation of Flap Flexibility of β-secretase Using Molecular Dynamic Simulations. Journal of Biomolecular Structure and Dynamics [Online] , 2015, 1-12. http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831
- ↑ 9.0 9.1 9.2 Golub, M. (Producer) & Golub, M. (Director). (2015). Youtube. United States: University of Rochester Introductory Biochemistry
- ↑ 10.0 10.1 Manada, N., Tanokashira, D., Hoska, A., Kametani, F., Tamoka, A,. and Araki, W. (2015). Amyloid beta-protein oligomers upregulate the beta-secretase, BACE1, through a post-translational mechanism involving its altered subcellular distribution in neurons. Molecular Brain, Vol. 8, p.1-12. 12p. doi: 10.1186/s13041-015-0163-5
- ↑ Sugana, K., Padlan E., Smith, C., Carlson, W., and Davis, D. (1987). Retrieved April 5, 2016 from

- ↑ JCSciphile, March - Beta-Secretase, 2014, Web, Retrieved April 5, 2016 from http://jcsciphile.com/molecule-of-the-month/march-beta-secretase/
- ↑ Vasser, R. The B-Secretase Enzyme in Alzheimer’s Disease. Journal of Molecular Neurosceince. 2004, 23, 105-113. http://link.springer.com/article/10.1385/JMN:23:1-2:105
- ↑ Shoji, M.; Golde, T.; Ghiso, J.; Cheung, T.; Estus, S.; Shaffer, L.; Cai, X.; Mckay, D.; Tintner, R.; Frangione, B.; Et, A. Production Of the Alzheimer Amyloid Beta Protein by Normal Proteolytic Processing. Science. 1992, 258, 126–129.
- ↑ Thompson, L.; Bronson, J.; Zusi, C.; Progress in the discovery of BACE Inhibitors. Current Pharmaceutical design. 2005, 11, 3383-3404.