Beta Secretase (BACE1) 1SGZ
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
</ref>. | </ref>. | ||
| - | Another notable structure found in beta-secretase is the S3 pocket. The S3 pocket is adjacent to the Thr 232 and the amino acids from the 10s loop, which contains Glycine and Glutamine. The Glycine located in the 10s loop forms a hydrogen bond with the substrate that helps to stabilize the 10s loop. This further balances the interaction of the beta-secretase substrate. In an open conformation of the 10s loop, the substrate and the S3 pocket have a greater affinity to bind with each other. The stabilization of the structure depends on the active site, flap and 10s loop <ref> Kumalo, M.; Soumendranath B.; Soliman, M. E. Investigation of Flap Flexibility of β-secretase Using Molecular Dynamic Simulations. Journal of Biomolecular Structure and Dynamics [Online] , 2015, 1-12. | + | Another notable structure found in beta-secretase is the S3 pocket. The S3 pocket is adjacent to the <scene name='72/728011/Thr232/1'>Thr 232</scene> and the amino acids from the 10s loop, which contains Glycine and Glutamine. The Glycine located in the 10s loop forms a hydrogen bond with the substrate that helps to stabilize the 10s loop. This further balances the interaction of the beta-secretase substrate. In an open conformation of the 10s loop, the substrate and the S3 pocket have a greater affinity to bind with each other. The stabilization of the structure depends on the active site, flap and 10s loop <ref> Kumalo, M.; Soumendranath B.; Soliman, M. E. Investigation of Flap Flexibility of β-secretase Using Molecular Dynamic Simulations. Journal of Biomolecular Structure and Dynamics [Online] , 2015, 1-12. |
http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831</ref>. | http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831</ref>. | ||
Revision as of 15:46, 21 April 2016
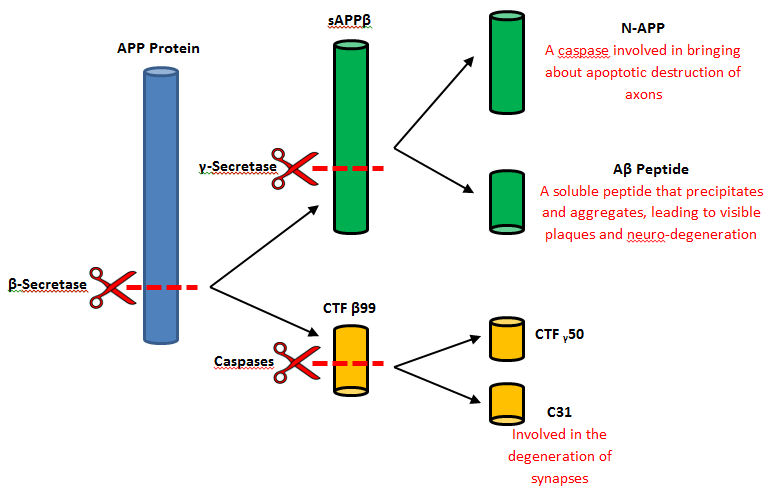
Beta-secretase, also known as BACE or Memapsin 2, is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing amyloid precursor protein (APP), which is an integral membrane protein.[1] A malfunction in the processing of APP results in the formation of the peptide amyloid beta. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with Alzheimers Disease. [2] Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.[3]
| |||||||||||
References
- ↑ Ghosh, A.; Kumaragurubaran, N.; Hong, L.; Koelsh, G.; Tang, J. Memapsin 2 (Beta-Secretase) Inhibitors: Drug Development. CAR Current Alzheimer Research. [Online] 2008, 5, 121–131. http://jamesmadisonva.library.ingentaconnect.com/content/ben/car/2008/00000005/00000002/art00004
- ↑ 2.0 2.1 2.2 John, V.; Beck, J. P.; Bienkowski, M. J.; Sinha, S.; Heinrikson, R. L. Human β-Secretase (BACE) And BACE Inhibitors. ChemInform. [Online] 2004, 35, 4625-4630. http://pubs.acs.org/doi/pdf/10.1021/jm030247h
- ↑ Hu, X.; Hicks, C. W.; He, W.; Wong, P.; Macklin, W. B.; Trapp, B. D.; Yan, R. Bace1 Modulates Myelination in the Central and Peripheral Nervous System. Nature Neuroscience Nat Neurosci. [Online] 2006, 9, 1520–1525. http://www.nature.com/neuro/journal/v9/n12/abs/nn1797.html
- ↑ Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. The Enzyme List Class 3 — Hydrolases. http://www.enzyme-database.org/downloads/ec3.pdf
- ↑ DBGET Search. KEGG ENZYME: 3.4.23.46. http://www.genome.jp/dbget-bin/www_bget?ec:3.4.23.46
- ↑ 6.0 6.1 6.2 Venugopal, C.; Demos, C. M.; Rao, K. S. J.; Pappolla, M. A.; Sambamurti, K. Beta-secretase: Structure, Function, and Evolution. CNS & Neurological Disorders Drug Targets. [Online] 2008. 7(3), 278–294. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2921875/
- ↑ 7.0 7.1 7.2 Shimizu, H.; Tosaki, A.; Kaneko, K.; Hisano, T.; Sakurai, T.; Nukina, N. Crystal Structure of an Active Form of BACE1, an Enzyme Responsible for Amyloid Beta Protein Production. Molecular and Cellular Biology [Online] 2008, 28(11), 3663-671. http://mcb.asm.org/content/28/11/3663.full.pdf+html
- ↑ Kumalo, M.; Soumendranath B.; Soliman, M. E. Investigation of Flap Flexibility of β-secretase Using Molecular Dynamic Simulations. Journal of Biomolecular Structure and Dynamics [Online] , 2015, 1-12. http://www.tandfonline.com/doi/full/10.1080/07391102.2015.1064831
- ↑ 9.0 9.1 9.2 Golub, M. (Producer) & Golub, M. (Director). (2015). Youtube. United States: University of Rochester Introductory Biochemistry
- ↑ 10.0 10.1 Manada, N., Tanokashira, D., Hoska, A., Kametani, F., Tamoka, A,. and Araki, W. (2015). Amyloid beta-protein oligomers upregulate the beta-secretase, BACE1, through a post-translational mechanism involving its altered subcellular distribution in neurons. Molecular Brain, Vol. 8, p.1-12. 12p. doi: 10.1186/s13041-015-0163-5
- ↑ Sugana, K., Padlan E., Smith, C., Carlson, W., and Davis, D. (1987). Retrieved April 5, 2016 from

- ↑ JCSciphile, March - Beta-Secretase, 2014, Web, Retrieved April 5, 2016 from http://jcsciphile.com/molecule-of-the-month/march-beta-secretase/
- ↑ Vasser, R. The B-Secretase Enzyme in Alzheimer’s Disease. Journal of Molecular Neurosceince. 2004, 23, 105-113. http://link.springer.com/article/10.1385/JMN:23:1-2:105
- ↑ Shoji, M.; Golde, T.; Ghiso, J.; Cheung, T.; Estus, S.; Shaffer, L.; Cai, X.; Mckay, D.; Tintner, R.; Frangione, B.; Et, A. Production Of the Alzheimer Amyloid-beta Protein by Normal Proteolytic Processing. Science. 1992, 258, 126–129.
- ↑ AnaSpec. Amyloid Peptides. Peptides, http://www.anaspec.com/products/productcategory.asp?id=1
- ↑ Thompson, L.; Bronson, J.; Zusi, C.; Progress in the discovery of BACE Inhibitors. Current Pharmaceutical design. 2005, 11, 3383-3404
- ↑ 17.0 17.1 Ghosh, A. K.; Brindisi, M.; Tang, J. Developing β-Secretase Inhibitors for Treatment of Alzheimer’s Disease. Journal of Neurochemistry. [Online] 2011, 120, 71–83.http://onlinelibrary.wiley.com/doi/10.1111/j.1471-4159.2011.07476.x/full