This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
CRISPR-Cas9
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
===Transcriptional regulation with CRISPR-Cas9=== | ===Transcriptional regulation with CRISPR-Cas9=== | ||
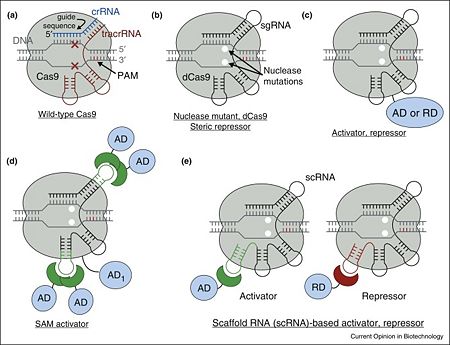
| + | Cas9 is a key protein of bacterial Type II CRISPR adaptive immune system. In its native context, Cas9 is an RNA-guided endonuclease that is responsible for targeted degradation of the invading foreign DNA–plasmids and phages. Cas9 is directed to its DNA targets by forming a ribonucleoprotein complex with two small non-coding RNAs: CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) (Figure 1a). By elegant engineering, crRNA and tracrRNA can be joined end-to-end and transcribed as a single guide RNA (sgRNA) that too efficiently directs Cas9 protein to DNA targets encoded within the guide sequence of sgRNA <ref name="Jinek">PMID:22745249</ref>. The optimal DNA target of the complex is determined by a Watson–Crick base pairing of a short ∼20-nt sequence within sgRNA (within the crRNA in wild-type), termed the guide sequence, adjacent to a few nucleotide long conserved motif recognized directly by Cas9 protein (protospacer adjacent motif, PAM) <ref name="Jinek">PMID:22745249</ref>. Despite this, a few mismatches between guide sequence and target DNA can be tolerated <ref name="Jinek">PMID:22745249</ref>, more so within the 5’ proximal position of the guide sequence. Cas9 nuclease can be converted into deactivated Cas9 (dCas9), an RNA-programmable DNA-binding protein, by mutating two key residues within its nuclease domains (Figure 1b) <ref name="Did">PMID:27344519</ref><ref name="Jinek">PMID:22745249</ref>. | ||
[[Image:1-s2.0-S0958166916301537-gr1.jpg|left|450px|thumb|'''Figure 1.''' Overview of Cas9 nuclease and dCas9-based transcription factors. (a) Wild-type Cas9 endonuclease guided by crRNA:tracrRNA to a specific site in DNA creates a double-stranded DNA break. (b) dCas9, nuclease deactivated mutant of Cas9, is an RNA programmable DNA binding protein. It can act as a steric repressor of transcription in prokaryotes and eukaryotes. sgRNA is an artificial chimeric molecule consisting of crRNA and tracrRNA molecules connected with a short loop. (c) dCas9 fusion with various transcription effectors can be used to repress or activate transcription. (d) Effector domains can be recruited by sgRNA in addition to dCas9 for enhanced activity. (e) sgRNA can be modified with specific protein binding hairpins to concurrently recruit repressor or activator domains in the same cell.<ref name="Did">PMID:27344519</ref>]] | [[Image:1-s2.0-S0958166916301537-gr1.jpg|left|450px|thumb|'''Figure 1.''' Overview of Cas9 nuclease and dCas9-based transcription factors. (a) Wild-type Cas9 endonuclease guided by crRNA:tracrRNA to a specific site in DNA creates a double-stranded DNA break. (b) dCas9, nuclease deactivated mutant of Cas9, is an RNA programmable DNA binding protein. It can act as a steric repressor of transcription in prokaryotes and eukaryotes. sgRNA is an artificial chimeric molecule consisting of crRNA and tracrRNA molecules connected with a short loop. (c) dCas9 fusion with various transcription effectors can be used to repress or activate transcription. (d) Effector domains can be recruited by sgRNA in addition to dCas9 for enhanced activity. (e) sgRNA can be modified with specific protein binding hairpins to concurrently recruit repressor or activator domains in the same cell.<ref name="Did">PMID:27344519</ref>]] | ||
| + | {{Clear}} | ||
==Crystal structure of a CRISPR RNA-guided surveillance complex, Cascade, bound to a ssDNA target<ref>PMID:25123481</ref>== | ==Crystal structure of a CRISPR RNA-guided surveillance complex, Cascade, bound to a ssDNA target<ref>PMID:25123481</ref>== | ||
Revision as of 10:53, 28 September 2016
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Didovyk A, Borek B, Tsimring L, Hasty J. Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr Opin Biotechnol. 2016 Aug;40:177-84. doi: 10.1016/j.copbio.2016.06.003. Epub, 2016 Jun 23. PMID:27344519 doi:http://dx.doi.org/10.1016/j.copbio.2016.06.003
- ↑ 2.0 2.1 2.2 2.3 Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21. doi: 10.1126/science.1225829. Epub 2012, Jun 28. PMID:22745249 doi:http://dx.doi.org/10.1126/science.1225829
- ↑ Mulepati S, Heroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014 Aug 14. pii: 1256996. PMID:25123481 doi:http://dx.doi.org/10.1126/science.1256996
- ↑ Hayes RP, Xiao Y, Ding F, van Erp PB, Rajashankar K, Bailey S, Wiedenheft B, Ke A. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016 Feb 25;530(7591):499-503. doi: 10.1038/nature16995. Epub 2016 Feb, 10. PMID:26863189 doi:http://dx.doi.org/10.1038/nature16995