Sandbox reserved 1225
From Proteopedia
| Line 24: | Line 24: | ||
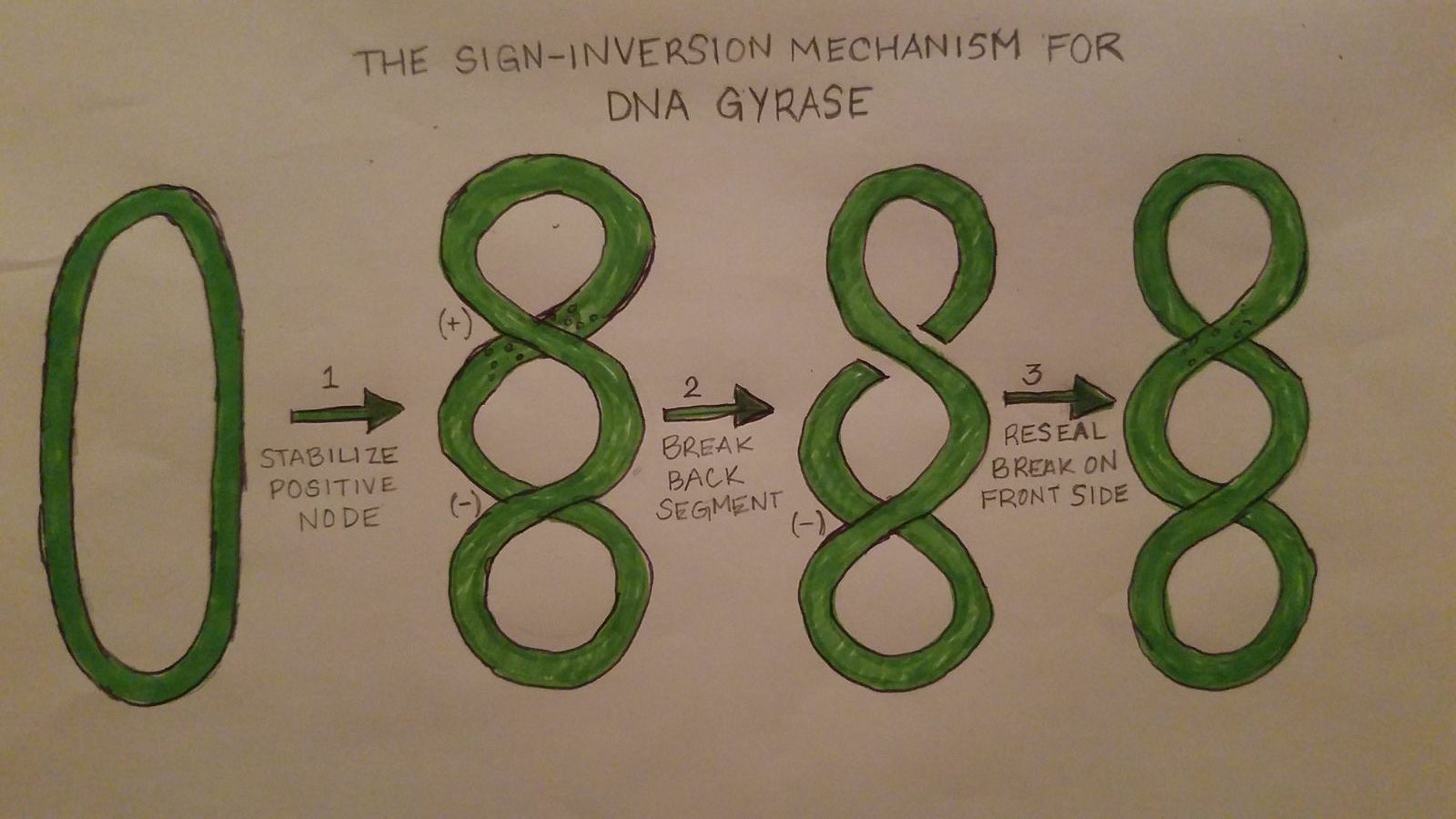
DNA Gyrase works by introducing conformational changes to itself as well as the protein complex. The Gyrase enzyme begins by interacting with the DNA. This causes the formation of the three gates that can be opened and closed. The G-segment, introduced under the “structure” section, forms a bond and binds to the first of the 3 gates, the central DNA gate. This binding of the gate produces chiral wrapping which in then produces a T-segment with in the last of the 3 gates, the N gate. After which, the N-gate binds with ATP which then causes the gate to close. Once this process is completed, the G-segment is released from the last of the 3 gates, the C-gate. This process causes a net result of 2 negative supercoils and a linkage difference of 2 from the initial linkage. | DNA Gyrase works by introducing conformational changes to itself as well as the protein complex. The Gyrase enzyme begins by interacting with the DNA. This causes the formation of the three gates that can be opened and closed. The G-segment, introduced under the “structure” section, forms a bond and binds to the first of the 3 gates, the central DNA gate. This binding of the gate produces chiral wrapping which in then produces a T-segment with in the last of the 3 gates, the N gate. After which, the N-gate binds with ATP which then causes the gate to close. Once this process is completed, the G-segment is released from the last of the 3 gates, the C-gate. This process causes a net result of 2 negative supercoils and a linkage difference of 2 from the initial linkage. | ||
| + | |||
| + | [[Image:Gyrase Function.jpg]] | ||
== '''Disease''' == | == '''Disease''' == | ||
Revision as of 15:32, 5 May 2017
DNA Gyrase
| |||||||||||
== References == Schoeffler, A. J., May, A. P., & Berger, J. M. (2010). A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Research, 38(21), 7830-7844. doi:10.1093/nar/gkq665
Basu, Aakash, et al. "Structural Dynamics and Mechanochemical Coupling in DNA Gyrase." Journal of Molecular Biology, vol. 428, no. Part B, 08 May 2016, pp. 1833-1845. EBSCOhost, doi:10.1016/j.jmb.2016.03.016.
Biochemistries. "The Mechanism of Negative DNA Supercoiling A..." Biochemistries. N.p., 23 Feb. 2014. Web. 04 May 2017.
Database, GeneCards Human Gene. "TOP2A Gene(Protein Coding)." GeneCards Is a Searchable, Integrative Database That Provides Comprehensive, User-friendly Information on All Annotated and Predicted Human Genes. N.p., n.d. Web. 04 May 2017
"DNA Gyrase." DNA Gyrase - ScienceDirect Topics. N.p., n.d. Web. 04 May 2017.
Gore, J., Z. Bryant, M. D. Stone, M. Nöllmann, N. R. Cozzarelli, and C. Bustamante. "Mechanochemical Analysis of DNA Gyrase Using Rotor Bead Tracking." Nature. U.S. National Library of Medicine, 05 Jan. 2006. Web. 04 May 2017.
Rahimi, H., Najafi, A., Eslami, H., Negahdari, B., & Moghaddam, M. M. (2016). Identification of novel bacterial DNA gyrase inhibitors: An in silico study. Journal Of Research In Pharmaceutical Sciences, 11(3), 250-258.
Schoeffler, A. J., May, A. P., & Berger, J. M. (2010). A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Research, 38(21), 7830-7844. doi:10.1093/nar/gkq665
Travers, A., & Muskhelishvili, G. (2015). DNA structure and function. FEBS Journal, 282(12), 2279-2295. doi:10.1111/febs.13307
Weigel, L. M., G. J. Anderson, and F. C. Tenover. "DNA Gyrase and Topoisomerase IV Mutations Associated with Fluoroquinolone Resistance in Proteus Mirabilis."Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Aug. 2002. Web. 04 May 2017.
Wendorff, T.J, B.H Schmidt, P. Heslop, C.A Austin, and J.M. Berger. "The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage." J.Mol.Biol. N.p., n.d. Web. 04 May 2017.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644