UDP-galactopyranose mutase, UDP-D-Galactopyranose furanomutase[1] or flavoenzyme uridine 5′-diphosphate galactopyranose mutase (UGM)[2].
It has a registry number of EC 5.4.99.9[1][3]; and is an isomerase, (L) polypeptide that consists of a dimer made up of 2 monomers, a homodimer[4][5][6].
Each monomer consists of a single chain of 390 amino acid residues, and 2 domains each: a GLF (UDP-galactopyronase mutase family) and a DAO (FAD dependent oxidoreductase domain)[4][5][6][2].
Each chain is 33% helical (17 helices; 131 residues) and 20% beta sheet (19strands; 81 residues)[4].
UDP-galactopryranose mutase consists of the genes glf and rfbd isolated from Klebisiella pneumoniae (stain 01 (ATCC 13882)) and Escherichia coli (stain BI21(de3)) through a plasma vector[4]; can also be found in Mycobacteria tuberculosis[2][5][6][7][8].
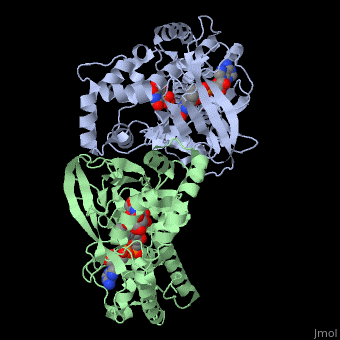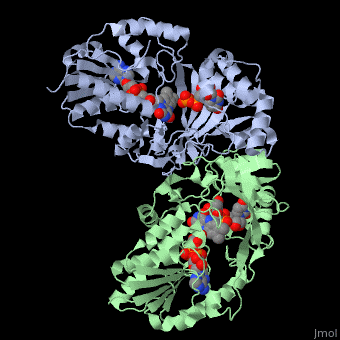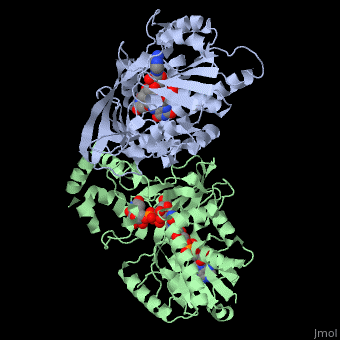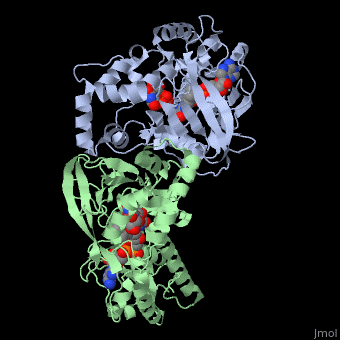
This is the a close up view of the of one of one of the monomers that make up UDP-galactopryranose mutase.
- , monomer A. Water molecules shown as red spheres.
History
The structure of the enzyme, UDP-galactopryranose mutase, was first solved by X-ray diffraction, at a resolution of 2.51Å, by a crystalline structure up tained through vapour diffusion by a hanging drop process, at a pH of 5.6 and temperature of 289.0K[4].
The enzyme-substrate complex structure was first crystallized to a 2.5Å resolution using the substrate analogue UDP-glucose (UDP-Glc) that binds the same at the same active site as the native substrate but does not react to give product due to having an equatorial C4-OH group rather than an axial one[6]; meaning that the UPD-Glc’s C4-OH group can not engage in a hydrogen bond with C4 carbonyl of the reduced flavin cofactor that the native substrate’s, Galactose’s, C4-Oh group can[2].
Structures related to, similar to, the structure of UDP-galactopryranose (3int) include those of: 3int, 1v0j, 1wam, 2bi7, 2bi8, 3gf4, 3inr[4].
Reaction
UDP-galactopryranose mutase catalyses the interconvertion of UPD-Galactopyranose, UDP-D-Galactopyranose, (UDP-Galp) to, UDP D Galacto-1-4-Furanose, (UPD-Galf) with the help of noncovalentely bound, reduced flavoprotien, (dihydro)flavin adenine dinucleotide (FAD)[1][2][3][4][5][6][7][8]with galactose-uridine-5’-diphosphate and uridine-5’-diphophate[4].
Reduction of the FAD involves a transformation of the coenzyme from a highly conjugated planar frame to a bent butterfly structure, which induces a conformational change within the enzyme making in more conductive to catalysis, and increases activity rate[2][7]. This flavin reduction also results in the translocation of the mobile loop inward toward the substrate with the Uridine portion of the ligand moving upward toward the flavin[2]. The flavin reduction and corresponding conformational change results in the enzyme being primed for covalent catalysis[2].
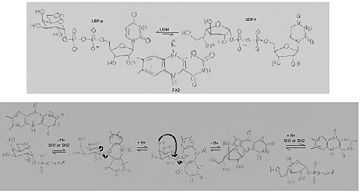
The inter-conversion of UDP-Gal
p and UPD-Gal
f catalyzed by UGM, through a nucleophilic S
N1 or S
N2 attack by reduced FAD
[6]The conversion between UDP-Galp and UPD-Galf involves the cleavage of the anomeric C-O bond, the reaction involves distortion of the ring allowing access to the O4 on C-1 to release the UDP[2][7].
There is a covalent adduct created between the flavin and the substrate that involves a nucleophilic attack (SN1/2) by the nitrogen atom (N5) of the isoalloxazine ring on the substrate which creates covalent falvin-galacrose adduct, that affords the formation of an iminium intermediate in the mechanism, and a displaces the UPD, as a good leaving group, which in turn allows the galactose ring to attach to the flavin, rearrange, dissociate and give the final product[2][5][6].
The C4 of the flavin provides a means to shuttle protons from the C4-OH to the nascent C5-OH after ring opening[2].
The substrate-binding, whose site is adjacent to the flavin cofactor directs the dynamic bridging of a recognition loop with distal FAD, which works to close the recognition loop over the substrate in the active site, and W160 sites, which interact with the Uralic of the substrate and is the N-terminal anchor of the recognition loop[6][8].
Arg174, located outside the putative active site on a mobile loop (recognition loop) adjacent to the putative active site pointing away from the flavin; once the substrate binds it closes this loop and brings the Arg174 side chain in toward the pyrophosphoryl group of the ligand[6]. This ‘recognition’ loop closes over the substrate-binding site, and is there by ‘locked’ down by the Arg174 coordination of the α-phosphate[2][8].
Binding sites and important residues

(A)This is an image of the important residues involved in binding in ribbon form; the orange is the recognition loop, while the green are various other residues described in the text to the right.
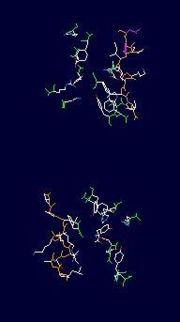
(B)This is an image of the important residues' sidechains involved in binding in stick form; the slighlty orange backbone is the recognition loop, while the slightly green backbones are various other residues described in the text to the left.
The cleft to which the flavin binds, in each monomer, is, adjacent to the binding site and is, lined with conserved residues 167-177: His63, Tyr155, Gln159, Trp160, Try185, Phe186, Arg250, Asn270, Arg280, Tyr314 and Asp351[2][5][6]. These, collectively, have a charge density consistent with binding of a negatively charged substrate such as reduced FAD[5][6][8].
Other residues involved in the conformational change of UGM, once the substrate is bound, are: Phe152, which sandwiches the Uralic ring of the ligand between it’s aromatic ring and that of Tyr155, and Leu175, which is located on the mobile loop moves to be adjacent to the bridging O of the ribose moiety and Phe152[6].
To position the sugar moiety the flavin, Tyr349, Arg280, Asn84, and an ordered water molecule all work together while theArg174 coordinates the α and β phosphates of the UDP, and with Try349 stabilizes the closed loop ‘locking’ it down[2][8].
Arg280 also works to coordinate the α and β phosphates[6][8]. Arg280 is located in the flavin-binding site[6].
This closing of the loop involves residues 167-177 which are adjacent to the Uridine diphosphoryl portion if the ligand and involves a extension of a short helix composed of residues 169-171 encompassing residues 72-174 moving Arg174 in toward the active site[2][6].
Also
The understanding of the structure of UDP-galactopryranose mutase and the mechanism with which it binds its substrates and catalyzes its reaction is of importance to pharmaceutical drug therapy because UPD-Galactosefuranose and UPD-Galactopyranose are found in many pathogens, in their surface constituents, cell wall glycoconjugates and in a vital component of arabinogalactan that connects peptidoglycan and mycolic acids in myobacteria cell walls in the lipoplysaccaride (LPS) O antigens of some Gram-negative bacteria; but are not found in human/mammal tissues so the this enzyme, UDP-galactopryranose mutase, that interconverts them can be safely inhibited slowing and preventing the growth of pathogenic microbes such as Escherichia coli, Mycobacteria tuberculosis, or Klebsiella pneumoniae[2][4][5][6][7][8].