Immunodeficiency virus protease
From Proteopedia
| Line 32: | Line 32: | ||
The current study reports on the apo crystal structure of the <scene name='Journal:JBSD:36/Cv/3'>South African HIV-1 subtype C protease (C-SA PR)</scene>. Structure of <scene name='Journal:JBSD:36/Cv/4'>unbound HIV-1 PR</scene> with the active site triplet (D25, T26 and G27) shown in ball-and-stick representation, <font color='magenta'><b>hinge region in magenta (residues 35–42 and 57–61)</b></font>, and <span style="color:cyan;background-color:black;font-weight:bold;">flap region (residues 46–54) in cyan</span>. The relevance of this study cannot be underestimated because South Africa is at the epicenter of the HIV/AIDS pandemic. A detailed understanding of the molecular interactions between the drug and its target is required if we are to improve the design of protease inhibitors (PIs). Our study indicated that the loss of a salt bridge between <scene name='Journal:JBSD:36/Cv/5'>residues E35 and R57</scene> at the hinge region affects the flap dynamics of the apo C-SA PR which may reduce the affinity and, therefore, the efficacy of the current protease inhibitors toward the C-SA PR (<span style="color:deeppink;background-color:black;font-weight:bold;">subtype C-SA PR is in deeppink</span>, [[3u71]] and <span style="color:yellow;background-color:black;font-weight:bold;">subtype B PR is in yellow</span>, [[2pc0]]). <scene name='Journal:JBSD:36/Cv/6'>Structural alignment</scene> of of the <span style="color:deeppink;background-color:black;font-weight:bold;">C-SA PR (deep pink</span>, PDB ID: [[3u71]]), <span style="color:yellow;background-color:black;font-weight:bold;">consensus subtype B PR (yellow</span>, PDB ID: [[2pc0]]), and <span style="color:wheat;background-color:black;font-weight:bold;">subtype B-MDR PR (color wheat</span>, PDB ID: [[1rp1]]) reveals that the PRs under investigation do not differ significantly. The crystal structure of the C-SA PR will serve as a foundation to improve the rational design of PIs which will have a greater impact on anti-retroviral chemotherapy in sub-Saharan Africa. | The current study reports on the apo crystal structure of the <scene name='Journal:JBSD:36/Cv/3'>South African HIV-1 subtype C protease (C-SA PR)</scene>. Structure of <scene name='Journal:JBSD:36/Cv/4'>unbound HIV-1 PR</scene> with the active site triplet (D25, T26 and G27) shown in ball-and-stick representation, <font color='magenta'><b>hinge region in magenta (residues 35–42 and 57–61)</b></font>, and <span style="color:cyan;background-color:black;font-weight:bold;">flap region (residues 46–54) in cyan</span>. The relevance of this study cannot be underestimated because South Africa is at the epicenter of the HIV/AIDS pandemic. A detailed understanding of the molecular interactions between the drug and its target is required if we are to improve the design of protease inhibitors (PIs). Our study indicated that the loss of a salt bridge between <scene name='Journal:JBSD:36/Cv/5'>residues E35 and R57</scene> at the hinge region affects the flap dynamics of the apo C-SA PR which may reduce the affinity and, therefore, the efficacy of the current protease inhibitors toward the C-SA PR (<span style="color:deeppink;background-color:black;font-weight:bold;">subtype C-SA PR is in deeppink</span>, [[3u71]] and <span style="color:yellow;background-color:black;font-weight:bold;">subtype B PR is in yellow</span>, [[2pc0]]). <scene name='Journal:JBSD:36/Cv/6'>Structural alignment</scene> of of the <span style="color:deeppink;background-color:black;font-weight:bold;">C-SA PR (deep pink</span>, PDB ID: [[3u71]]), <span style="color:yellow;background-color:black;font-weight:bold;">consensus subtype B PR (yellow</span>, PDB ID: [[2pc0]]), and <span style="color:wheat;background-color:black;font-weight:bold;">subtype B-MDR PR (color wheat</span>, PDB ID: [[1rp1]]) reveals that the PRs under investigation do not differ significantly. The crystal structure of the C-SA PR will serve as a foundation to improve the rational design of PIs which will have a greater impact on anti-retroviral chemotherapy in sub-Saharan Africa. | ||
| - | + | ||
| - | [[HIV Protease Inhibitor Pharmacokinetics]] | + | ==See Also== |
| - | [[HIV Protease Inhibitor Resistance Profile]] | + | *''Aids Before Protease Inhibitors'' and ''HIV Protease Inhibitors: A Breakthrough'' at [[Molecular Playground/HIV Protease Inhibitor|HIV Protease Inhibitor]]. |
| - | [[HIV Protease Resistance]] | + | *[[HIV Protease Inhibitor Pharmacokinetics]] |
| - | [[Viability of a drug-resistant HIV-1 protease mutant]] | + | *[[HIV Protease Inhibitor Resistance Profile]] |
| - | [[HIV and accessory proteins]] | + | *[[HIV Protease Resistance]] |
| - | [[Treatments:HIV Protease Inhibitor Pharmacokinetics References]] | + | *[[Viability of a drug-resistant HIV-1 protease mutant]] |
| - | [[Group:SMART:HIV-1 Subtype C Protease]] | + | *[[HIV and accessory proteins]] |
| - | [[Human Immunodeficiency Virus]] | + | *[[Treatments:HIV Protease Inhibitor Pharmacokinetics References]] |
| - | [[Virus protease]] | + | *[[Group:SMART:HIV-1 Subtype C Protease]] |
| + | *[[Human Immunodeficiency Virus]] | ||
| + | *[[Virus protease]] | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
</StructureSection> | </StructureSection> | ||
Revision as of 16:05, 23 October 2018
| |||||||||||
Contents |
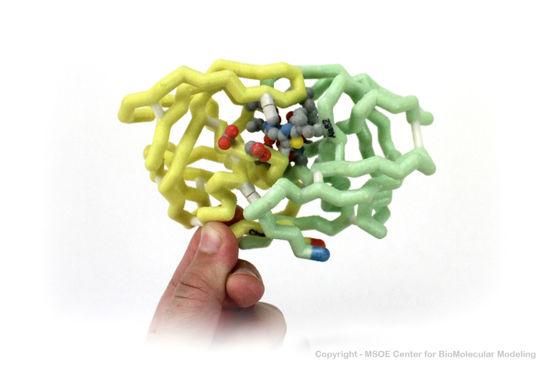
3D Printed Physical Model of HIV Protease
Shown below are 3D printed physical models of HIV Protease. Both versions are shown in alpha carbon format, with select side chains shown colored by element, with carbon gray, nitrogen blue, oxygen red and sulfur yellow. Both models have been designed with precisely embedded magnets that allow the two chains to pull apart into individual pieces.
The MSOE Center for BioMolecular Modeling
The MSOE Center for BioMolecular Modeling uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our Model Gallery.
3D structures of immunodeficiency protease
Updated on 23-October-2018
Additional Resources
For additional information, see:
- Human Immunodeficiency Virus
- Structural Biology of HIV, an interactive Flash graphic of the virion with explanations of its components.
References
- ↑ Tie Y, Wang YF, Boross PI, Chiu TY, Ghosh AK, Tozser J, Louis JM, Harrison RW, Weber IT. Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors. Protein Sci. 2012 Mar;21(3):339-50. doi: 10.1002/pro.2019. Epub 2012 Jan 24. PMID:22238126 doi:10.1002/pro.2019
- ↑ Wlodawer A, Miller M, Jaskolski M, Sathyanarayana BK, Baldwin E, Weber IT, Selk LM, Clawson L, Schneider J, Kent SB. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616-21. PMID:2548279
- ↑ Lapatto R, Blundell T, Hemmings A, Overington J, Wilderspin A, Wood S, Merson JR, Whittle PJ, Danley DE, Geoghegan KF, et al.. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989 Nov 16;342(6247):299-302. PMID:2682266 doi:http://dx.doi.org/10.1038/342299a0
- ↑ Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins. 2007 Apr 1;67(1):232-42. PMID:17243183 doi:10.1002/prot.21304
- ↑ Maschera B, Darby G, Palu G, Wright LL, Tisdale M, Myers R, Blair ED, Furfine ES. Human immunodeficiency virus. Mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J Biol Chem. 1996 Dec 27;271(52):33231-5. PMID:8969180
- ↑ Naicker P, Achilonu I, Fanucchi S, Fernandes M, Ibrahim MA, Dirr HW, Soliman ME, Sayed Y. Structural insights into the South African HIV-1 subtype C protease: impact of hinge region dynamics and flap flexibility in drug resistance. J Biomol Struct Dyn. 2012 Nov 12. PMID:23140382 doi:10.1080/07391102.2012.736774
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Michal Harel, Eran Hodis, Mark Hoelzer, David Canner, Eric Martz, Ann Taylor, Wayne Decatur, Alexander Berchansky, Jaime Prilusky, Karsten Theis