This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1471
From Proteopedia
| Line 19: | Line 19: | ||
<scene name='80/800650/Human_cox-2_bound_to_vioxx/3'>Rofecoxib</scene> | <scene name='80/800650/Human_cox-2_bound_to_vioxx/3'>Rofecoxib</scene> | ||
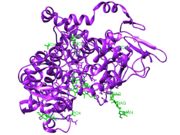
| - | [[Image: | + | [[Image:Heteroatoms of COX2.jpg|thumb|Figure 1: Ligands (green) bound to COX-2 from PDB file 5KIR.]] |
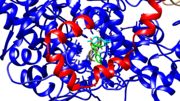
| - | [[Image:COX2 channel.jpeg|thumb|Figure | + | [[Image:COX2 channel.jpeg|thumb|Figure 2: Channel of the cyclooxygenase bound to Viox from PDB file 5KIR.]] |
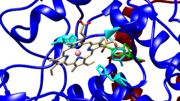
| - | [[Image:Peroxidase activity site.jpg|thumb|Figure | + | [[Image:Peroxidase activity site.jpg|thumb|Figure 3: Peroxidase activity site from PDB file 5KIR.]] |
===Domains=== | ===Domains=== | ||
Revision as of 22:45, 4 December 2018
| This Sandbox is Reserved from November 5 2018 through January 1, 2019 for use in the course "CHEM 4923: Senior Project taught by Christina R. Bourne at the University of Oklahoma, Norman, USA. This reservation includes Sandbox Reserved 1471 through Sandbox Reserved 1478. |
To get started:
More help: Help:Editing |
This page is reserved for Katie
Contents |
Cyclooxygenase 2
|
A Cyclooxygenase is an enzyme that catalyzes the transformation of arachidonic acid into prostaglandins, prostacyclins, and thromboxanes.[1] Another name for cyclooxygenase is prostaglandin H2 synthase. [2] There are two names because there are two catalytic activities: cyclooxygenase and peroxidase. The abbreviation for cyclooxygenase and prostaglandin H2 synthase is COX and PGHS respectively and can be used interchangeably. There are two types of cyclooxygenases: COX-1 and COX-2. COX-1 is responsible for platelet aggregation and gastric acidity.[1] COX-2 is involved in pathways that lead to inflammation (swelling), pain, and fever.[1]
Function
Reaction
Cyclooxygenases catalyzes arachidonic acid or other fatty acids into prostaglandin H2 (PGH2) and other molecules that can be used for signal transduction. Of the two catalytic activities, the cyclooxygenase reaction happens before the peroxidase reaction. However, the peroxidase activity activates the cyclooxygenase activity.[3] “Two-electron reduction of a peroxide substrate results in the oxidation of the ferric heme to an oxo-ferryl porphyrin radical cation.”[4] The most important catalytic residue is Tyrosine 385. It transfers and electron to the heme to create the radical on the tyrosine [4] The tyrosine then takes the pro-S hydrogen from carbon 13 of arachidonic acid to produce a radical on the arachidonic intermediate. [5] Two oxygen molecules are inserted to cyclize the intermediate.[5] Tyrosine 385 is reduced from the “peroxyl radical to the hyperoxide to form PGG2.”[4] PGG2 is then reduced by the peroxidase activity to form PGH2. (Lehninger). The Tyrosine 385 radical is regenerated, so the cyclooxygenase activity does not need to be activated for every reaction.[4]
COX-1
COX-2
Structure
Domains
COX-1 is made up of 576 amino acids and COX-2 is made up of 581 amino acids.[4] The sequences match with 60% identical amino acids.[4] The structures of these isozymes are nearly superimposable with three domains each. The made up of residues 73-116, goes through half of the lipid bilayer.[3] These residues are hydrophobic because they are in a hydrophobic environment with the fatty acid tails of the lipids surrounding this domain. Residues 117-587 make up the .[3] The catalytic domain has two catalytic active sites for each of the catalytic activities of cyclooxygenase and peroxidase. The third domain is similar to an .[3] The function of the EGF domain is not fully understood, but it is speculated to help with structure stability and protein-protein interactions.[3] This domain is linked to the catalytic domain by disulfide bridges between Cys 37- Cys 159.[3]
Heteroatoms of the PDB file 5KIR[1]
| Heteroatom Name | ID |
|---|---|
| Phosphate Ion | PO4 |
| Rofecoxib | RCX |
| Glycerol | GOL |
| Alpha-D-Mannose | MAN |
| Protoprophyrin IX Containing Cobalt | COH |
| Ammonium Ion | NH4 |
| N-Acetyl-D-Glucosamine | NAG |
Energetics
Medical Applications
References
- ↑ 1.0 1.1 1.2 1.3 Orlando BJ, Malkowski MG. Crystal structure of rofecoxib bound to human cyclooxygenase-2. Acta Crystallogr F Struct Biol Commun. 2016 Oct 1;72(Pt 10):772-776. Epub 2016, Sep 22. PMID:27710942 doi:http://dx.doi.org/10.1107/S2053230X16014230
- ↑ Smith, W. L and R. Langenbach (2001). "Why there are two cyclooxygenase isozymes." The Journal of Clinical Investigation 107(12):1491-1495.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Picot D, Loll PJ, Garavito RM. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature. 1994 Jan 20;367(6460):243-9. PMID:8121489 doi:http://dx.doi.org/10.1038/367243a0
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Rouzer CA, Marnett LJ. Cyclooxygenases: structural and functional insights. J Lipid Res. 2009 Apr;50 Suppl:S29-34. doi: 10.1194/jlr.R800042-JLR200. Epub 2008, Oct 23. PMID:18952571 doi:http://dx.doi.org/10.1194/jlr.R800042-JLR200
- ↑ 5.0 5.1 "Chapter 21: Lipid Biosynthesis." Lehninger Principles of Biochemistry, by David L. Nelson et al., Basingstoke, 2017, pp. 824-825.