We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1476
From Proteopedia
(Difference between revisions)
m (Addition to Disease) |
m (Change to title) |
||
| Line 18: | Line 18: | ||
| - | == Function == | + | == Function and Energetics == |
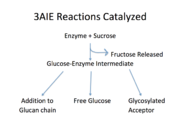
The major functions of this enzyme include synthesis of glucan, a glucosylated acceptor molecule, or release of free glucose.<ref name="Monchois">PMID:10234842</ref> [[Image:Reaction GSase.png|thumb|Reactions catalyzed by 3aie, mutansucrase of S. mutans. Image adapted from Monchois, et al.]] As shown in Figure 1, all of the products are released from the same active site and from a glucosyl-enzyme intermediate.<ref name="Monchois" /> The sole, natural substrate for this enzyme is sucrose.<ref name="Hamada">PMID:6446023</ref> No cofactors or energy-carriers are required for these reactions.<ref name="Monchois" /> The energy required for bond formation is solely provided by the energy released from sucrose hydrolysis.<ref name="Monchois" />This particular glucansucrase is a mutansucrase, meaning it catalyzes the formation of α(1,3) linked glucose moieties.<ref name="Ito" /> The latest research proposes that the glucose moieties are most likely added to the non-reducing end of the glucan chain.<ref name="Moulis">PMID:16864576</ref> Binding of a glucan chain to the active site may cause a conformational change within the enzyme that favors elongation of the glucan chain instead of the formation of the other products.<ref name="Monchois" /> Acceptor molecules include maltose and isomaltose.<ref name="Leemhuis" /> Free glucose may be released upon hydrolysis of the glucose-enzyme intermediate.<ref name="Leemhuis" /> | The major functions of this enzyme include synthesis of glucan, a glucosylated acceptor molecule, or release of free glucose.<ref name="Monchois">PMID:10234842</ref> [[Image:Reaction GSase.png|thumb|Reactions catalyzed by 3aie, mutansucrase of S. mutans. Image adapted from Monchois, et al.]] As shown in Figure 1, all of the products are released from the same active site and from a glucosyl-enzyme intermediate.<ref name="Monchois" /> The sole, natural substrate for this enzyme is sucrose.<ref name="Hamada">PMID:6446023</ref> No cofactors or energy-carriers are required for these reactions.<ref name="Monchois" /> The energy required for bond formation is solely provided by the energy released from sucrose hydrolysis.<ref name="Monchois" />This particular glucansucrase is a mutansucrase, meaning it catalyzes the formation of α(1,3) linked glucose moieties.<ref name="Ito" /> The latest research proposes that the glucose moieties are most likely added to the non-reducing end of the glucan chain.<ref name="Moulis">PMID:16864576</ref> Binding of a glucan chain to the active site may cause a conformational change within the enzyme that favors elongation of the glucan chain instead of the formation of the other products.<ref name="Monchois" /> Acceptor molecules include maltose and isomaltose.<ref name="Leemhuis" /> Free glucose may be released upon hydrolysis of the glucose-enzyme intermediate.<ref name="Leemhuis" /> | ||
Revision as of 17:50, 8 December 2018
| This Sandbox is Reserved from November 5 2018 through January 1, 2019 for use in the course "CHEM 4923: Senior Project taught by Christina R. Bourne at the University of Oklahoma, Norman, USA. This reservation includes Sandbox Reserved 1471 through Sandbox Reserved 1478. |
To get started:
More help: Help:Editing |
PDB ID 3AIE
This enzyme is one type of glucansucrase from Streptococcus mutans.[1] These enzymes are large and extracellularly expressed by few other bacterial species.[2] Their main function is to catalyze the production of glucose polymers from sucrose substrates.[1] These polymers comprise the dental plaque that promotes tooth decay in humans. For this reason, knowledge of the structure and function of this enzyme is critical for the development of an inhibitor in an effort to reduce the prevalence of cavities and periodontal disease.
| |||||||||||