Sandbox Reserved 1506
From Proteopedia
| Line 1: | Line 1: | ||
| - | <Structure load=' | + | <Structure load='Insert PDB code or filename here' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /><scene name='80/802680/Anaat/1'>ANAAT en couleur</scene> |
'<Structure load='1b6b' size='350' frame='true' align='right' caption='Protein Display ' scene='Insert optional scene name here' />'{{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | '<Structure load='1b6b' size='350' frame='true' align='right' caption='Protein Display ' scene='Insert optional scene name here' />'{{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
== Structure == | == Structure == | ||
| Line 7: | Line 7: | ||
== Function == | == Function == | ||
| + | Serotonin N-acetyltransferase is also named aralkylamine-N-acetyltransferase (ANAAT). | ||
| + | [[Image:melationin synthesis.jpg]] | ||
== Regulation == | == Regulation == | ||
| Line 16: | Line 18: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | [1] The Structural Basis of Ordered Substrate Binding by Serotonin N-Acetyltransferase: Enzyme Complex at 1.8 A ˚ Resolution with a Bisubstrate Analog - Alison Burgess Hickman,M. A. A. Namboodiri, David C. Klein, and Fred Dyda - Cell, Vol. 97, 361–369, April 30, 1999. | ||
| + | |||
| + | [2] Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N -acetyltransferase mediated by phosphoserine-205 - Surajit Ganguly, Joan L. Weller, Anthony Ho, Philippe Chemineau, Benoit Malpaux, and David C. Klein - PNAS, January 25, 2005. | ||
| + | |||
| + | [3] | ||
| + | |||
<references/> | <references/> | ||
Revision as of 11:33, 2 January 2019
|
|
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Contents |
Structure
This is a default text for your page b6. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
Serotonin N-acetyltransferase is also named aralkylamine-N-acetyltransferase (ANAAT).
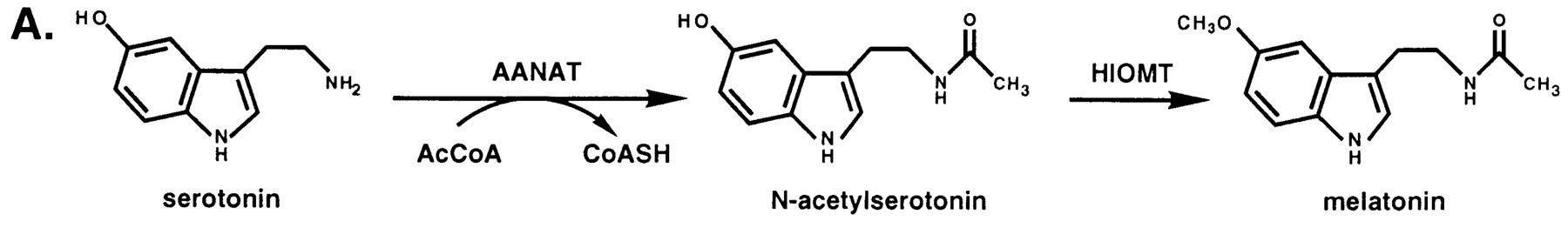
Regulation
Relevance
Structural highlights
</StructureSection>
References
[1] The Structural Basis of Ordered Substrate Binding by Serotonin N-Acetyltransferase: Enzyme Complex at 1.8 A ˚ Resolution with a Bisubstrate Analog - Alison Burgess Hickman,M. A. A. Namboodiri, David C. Klein, and Fred Dyda - Cell, Vol. 97, 361–369, April 30, 1999.
[2] Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N -acetyltransferase mediated by phosphoserine-205 - Surajit Ganguly, Joan L. Weller, Anthony Ho, Philippe Chemineau, Benoit Malpaux, and David C. Klein - PNAS, January 25, 2005.
[3]
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
