Sandbox Reserved 1506
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | <Structure load='1b6b' size='350' frame='true' align='right' caption='Protein Display ' scene='Insert optional scene name here' /> | |
| - | + | {{Sandbox_Reserved_ESBS}} | |
| Line 19: | Line 19: | ||
== Structure == | == Structure == | ||
| - | < | + | |
| + | '<Structure load='1b6b' size='350' frame='true' align='right' caption='Protein Display ' scene='ANAAT Structure' /> | ||
| + | |||
''' General description ''' | ''' General description ''' | ||
| - | + | <scene name='80/802680/Anaat/1'>ANAAT </scene> is a globular protein consisting of an 8-stranded β sheet with five α helices, with a conserved motif in the center of the sheet forming a binding site for acetyl-CoA, the acetyl source. | |
A long α helix lies on the concave surface of the β sheet, with its axis approximately aligned with those of the strands. Two other helices wraps around the convexe surface of the sheet, perpendicular to it, like the staves of a barrel. | A long α helix lies on the concave surface of the β sheet, with its axis approximately aligned with those of the strands. Two other helices wraps around the convexe surface of the sheet, perpendicular to it, like the staves of a barrel. | ||
Revision as of 20:58, 3 January 2019
|
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Contents |
Function
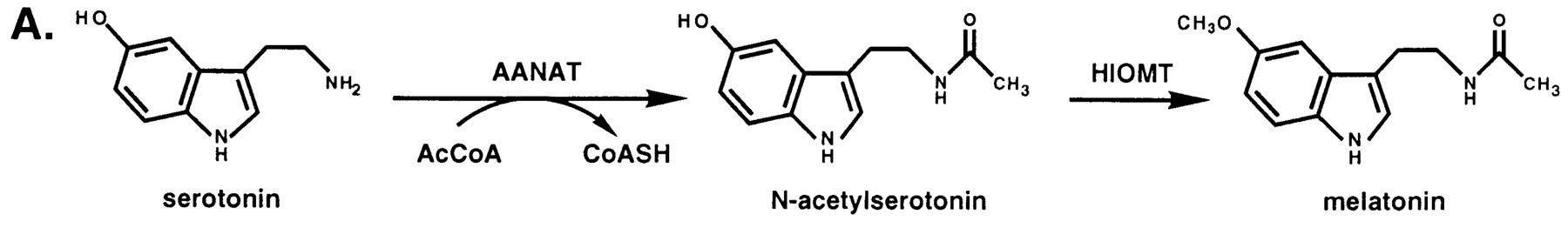
Serotonin N-acetyltransferase is also named aralkylamine-N-acetyltransferase (ANAAT). AANAT is a member of a large superfamily of proteins, referred to alternatively as the motif A/B or the GCN-5-related N-acetyltransferase (or GNAT) family. The role of these enzymes is to catalyse the acetylation all sorts of residues. ANAAT catalyzes the acetylation of the amine group on serotonin, an intermediate in melatonin synthesis. It is the penultimate enzyme in the melatonin pathway, as it is shown on fig1. [1].
Relevance
Circulating melatonin plays a role in the circadian rythm. Day/night differences in circulating melatonin levels provide a hormonal analog signal of environmental lighting, which is used in a variety of ways to optimize circadian and circannual rhythms in physiology[2]. The amount of circulating melatonin is correlated with ANAAT activity, as enzyme activity varies in parallel with melatonin amount. For example, in some species the night/day differences in melatonin, AANAT activity, and protein are 10- to 100-fold [3][4]. Defects in ANAAT regulation could be responsible for sleep troubles, such as insomnia or narcolepsy.[5] but also serotonin-related deseases such as depression [6].
Structure
'
|
General description
is a globular protein consisting of an 8-stranded β sheet with five α helices, with a conserved motif in the center of the sheet forming a binding site for acetyl-CoA, the acetyl source. A long α helix lies on the concave surface of the β sheet, with its axis approximately aligned with those of the strands. Two other helices wraps around the convexe surface of the sheet, perpendicular to it, like the staves of a barrel.
The Acetyl-CoA binding site
The pantetheine group of Ac-CoA binds in a cleft formed at the site where two parallel β strands come apart. It is held in place by several H-bonds. The turn joining a strand of the β sheet and the long α helix form a positively charged pocket to welcome the pyrophosphate group.
The active site
The folding of the active site is similar to the one of other members of the GNAT superfamily. However, the active site is deeply burried into the protein, which is rare in the GNAT family. The access to this site is warranted for serotonin by and hydrophobic funnel formed by three polypeptide loops which converge above the acCoA binding site. (INSERT GREEN LINK HERE WHICH SHOWS THE HYDROPHOBIC FUINNEL AND THE ACTIVE SITE)
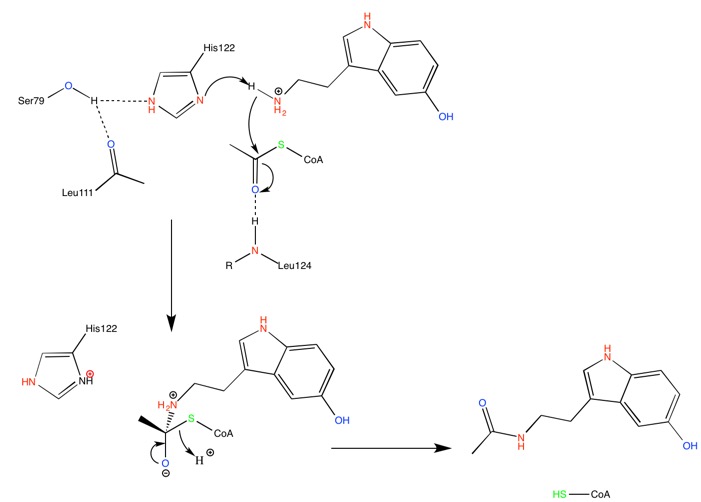
At the bottom of the funnel, in the active site, are located two conserved histidines (GREEN LINK HERE) which suggest a catalytic mechanism for acetylation involving imidazole groups acting as general acid/base catalysts [7]. The supposed calytic role of these histidines, H120 and H122, is illustrated by the following mechanism.
His-122 is hold in place through H-bond interaction with a serine residue, Ser-97, suggesting this to be the main catalytic residue. Mutations of either of these histidine residues result in a complete loss of catalytic function.
Right after catalytic histidine H122, Ala-123 is responsible for a bulge in the β sheet. It is an important feature of the active site, and it also accentuate the twist of the β sheet.
Access to the active site
One of the three loops forming the funnel is rather mobile. Many hydrophobic residues are lining the sides of the funnel
Regulation
Références
[1] Hickman, A. B; Klein, D. C; Dyda, F. Melatonin Biosynthesis: The Structure of Serotonin N-Acetyltransferase at 2.5A Resolution Suggests a Catalytic Mechanism. Mol. Cell. 1999, 3-1, 23-32.
[2] Arendt, J. (1995) Melatonin and the Mammalian Pineal Gland (Chapman & Hall, London), pp. 201–285.
[3] Klein, D.C., and Weller, J.L. (1972). Rapid light induced decrease in pineal serotonin N-acetyltransferase activity. Science 177, 532–533.
[4]Gastel, J.A., Roseboom, P.H., Rinaldi, P.A., Weller, J.L., and Klein,D.C. (1998). Melatonin production: proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science 279, 1358–1360.
[5] Rati Srinivasan, Guy Gechtman, Abe Bayer. What is the Molecular Basis of Sleep Regulation, https://sites.tufts.edu/sleep/
[6] Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N -acetyltransferase mediated by phosphoserine-205 - Surajit Ganguly, Joan L. Weller, Anthony Ho, Philippe Chemineau, Benoit Malpaux, and David C. Klein - PNAS, January 25, 2005.
[7] The Structural Basis of Ordered Substrate Binding by Serotonin N-Acetyltransferase: Enzyme Complex at 1.8 A ˚ Resolution with a Bisubstrate Analog - Alison Burgess Hickman,M. A. A. Namboodiri, David C. Klein, and Fred Dyda - Cell, Vol. 97, 361–369, April 30, 1999.
- ↑ Hickman, A. B; Klein, D. C; Dyda, F. Melatonin Biosynthesis: The Structure of Serotonin N-Acetyltransferase at 2.5A Resolution Suggests a Catalytic Mechanism. Mol. Cell. 1999, 3-1, 23-32
- ↑ Arendt, J. (1995) Melatonin and the Mammalian Pineal Gland (Chapman & Hall, London), pp. 201–285.
- ↑ Klein, D.C., and Weller, J.L. (1972). Rapid light induced decrease in pineal serotonin N-acetyltransferase activity. Science 177, 532–533.
- ↑ Gastel, J.A., Roseboom, P.H., Rinaldi, P.A., Weller, J.L., and Klein,D.C. (1998). Melatonin production: proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science 279, 1358–1360
- ↑ Rati Srinivasan, Guy Gechtman, Abe Bayer. What is the Molecular Basis of Sleep Regulation, https://sites.tufts.edu/sleep/
- ↑ Hickman, A. B; Klein, D. C; Dyda, F. Melatonin Biosynthesis: The Structure of Serotonin N-Acetyltransferase at 2.5A Resolution Suggests a Catalytic Mechanism. Mol. Cell. 1999, 3-1, 23-32
- ↑ Hickman, A. B; Klein, D. C; Dyda, F. Melatonin Biosynthesis: The Structure of Serotonin N-Acetyltransferase at 2.5A Resolution Suggests a Catalytic Mechanism. Mol. Cell. 1999, 3-1, 23-32.