User:Ashley Crotteau/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
==Structure== | ==Structure== | ||
| - | <scene name='81/811707/Kmt_overall_structure/2'>Overall Structure</scene> | ||
| - | <scene name='81/811707/Alpha_helices/1'>Alpha Helices</scene> | ||
| - | |||
| - | <scene name='81/811707/Active_site/1'>Active Site</scene> | ||
| - | |||
| - | Active Site<ref name="Xiao" /> | ||
| - | |||
| - | <scene name='81/811707/Kmt_active_site/2'>KMT Active Site Residues</scene> | ||
| - | |||
| - | ===SET 7/9=== | ||
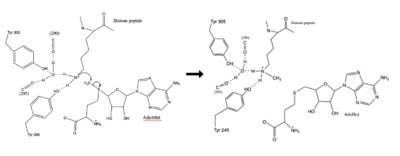
The human lysine methyltransferase (HKMT) SET7/9 is 366 amino acids long. The overall structure looks like a dimer although it acts as a monomer. The structure is composed of the ΔSET7/9 domain. The ΔSET7/9 consists of the SET domain along with the pre- and post-SET regions. The pre- and post-SET regions are adjacent to SET domain and are cysteine rich. The pre-SET cysteine region is located near the N-terminal where the post-SET region is located near the C-terminal of the domain. These regions are said to play an important role in substrate recognition and enzymatic activity.<ref name="Schubert" /><ref name="Yeates" /> | The human lysine methyltransferase (HKMT) SET7/9 is 366 amino acids long. The overall structure looks like a dimer although it acts as a monomer. The structure is composed of the ΔSET7/9 domain. The ΔSET7/9 consists of the SET domain along with the pre- and post-SET regions. The pre- and post-SET regions are adjacent to SET domain and are cysteine rich. The pre-SET cysteine region is located near the N-terminal where the post-SET region is located near the C-terminal of the domain. These regions are said to play an important role in substrate recognition and enzymatic activity.<ref name="Schubert" /><ref name="Yeates" /> | ||
Revision as of 23:06, 4 April 2019
H. sapiens Lysine Methyltransferase, SET 7/9
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003 Jun;28(6):329-35. PMID:12826405
- ↑ 2.0 2.1 2.2 Yeates TO. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002 Oct 4;111(1):5-7. PMID:12372294
- ↑ 3.0 3.1 3.2 3.3 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 4.0 4.1 Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998 Jun 26;273(26):15933-9. PMID:9632640
- ↑ 5.0 5.1 doi: https://dx.doi.org/10.1016/C2014-0-02189-2
- ↑ https://en.wikipedia.org/wiki/SET_domain#Structure