User:Ashley Crotteau/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
[https://en.wikipedia.org/wiki/Histone_methylation Histone Methylation] | [https://en.wikipedia.org/wiki/Histone_methylation Histone Methylation] | ||
==Structure== | ==Structure== | ||
| + | |||
| + | ===SET Domain=== | ||
The human lysine methyltransferase (HKMT) SET7/9 is 366 amino acids long. The overall structure looks like a dimer although it acts as a monomer. The structure is composed of the ΔSET7/9 domain. The ΔSET7/9 consists of the SET domain along with the pre- and post-SET regions. The pre- and post-SET regions are adjacent to SET domain and are cysteine rich. The pre-SET cysteine region is located near the N-terminal where the post-SET region is located near the C-terminal of the domain. These regions are said to play an important role in substrate recognition and enzymatic activity.<ref name="Schubert" /><ref name="Yeates" /> | The human lysine methyltransferase (HKMT) SET7/9 is 366 amino acids long. The overall structure looks like a dimer although it acts as a monomer. The structure is composed of the ΔSET7/9 domain. The ΔSET7/9 consists of the SET domain along with the pre- and post-SET regions. The pre- and post-SET regions are adjacent to SET domain and are cysteine rich. The pre-SET cysteine region is located near the N-terminal where the post-SET region is located near the C-terminal of the domain. These regions are said to play an important role in substrate recognition and enzymatic activity.<ref name="Schubert" /><ref name="Yeates" /> | ||
| - | The SET domain is mostly defined by <scene name='81/811707/Variable_knot/2'>turns and loops</scene> with the few <scene name='81/811707/Beta_sheets/3'>antiparallel β-sheets</scene>.<ref name="Schubert" /> <scene name='81/811707/Beta-hairpin/2'>Residues 337-349</scene> form a β-hairpin that sticks out at a right angle to the surface of the enzyme. The following three residues (<scene name='81/811707/Sharp_bend/2'>350-352</scene>) accommodate a sharp bend in the peptide chain and the end of the protein adopts an <scene name='81/811707/C-term_alpha_helix/2'>α-helical conformation</scene>.<ref name="Xiao" /> The two most defining features of the SET domain are the C-terminal tyrosine and the knot-like fold. These two components have been recognized to be essential for S-adenosyl-L-methionine (SAM) binding and catalysis.<ref name="Schubert" /> <ref name="Yeates" /> <ref name="Huang" /> The knot-like fold contains the binding sites for the cofactor SAM and the peptide substrate.<ref name="Licciardello" /> | + | The SET domain is mostly defined by <scene name='81/811707/Variable_knot/2'>turns and loops</scene> with the few <scene name='81/811707/Beta_sheets/3'>antiparallel β-sheets</scene>.<ref name="Schubert" /> <scene name='81/811707/Beta-hairpin/2'>Residues 337-349</scene> form a β-hairpin that sticks out at a right angle to the surface of the enzyme.<ref name="Xiao" /> The following three residues (<scene name='81/811707/Sharp_bend/2'>350-352</scene>) accommodate a sharp bend in the peptide chain and the end of the protein adopts an <scene name='81/811707/C-term_alpha_helix/2'>α-helical conformation</scene>.<ref name="Xiao" /> The two most defining features of the SET domain are the C-terminal tyrosine and the knot-like fold. These two components have been recognized to be essential for S-adenosyl-L-methionine (SAM) binding and catalysis.<ref name="Schubert" /> <ref name="Yeates" /> <ref name="Huang" /> The knot-like fold contains the binding sites for the cofactor SAM and the peptide substrate.<ref name="Licciardello" /> |
| - | + | ===Active Site and Channel=== | |
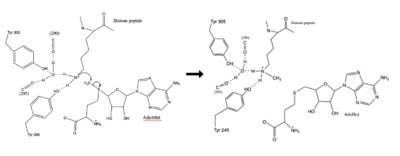
| - | The | + | The most notable feature of the HKMT is the presence of the lysine access channel as the active site. The cofactor and peptide substrate are actually located on opposite sides of the SET domain but are connected through this narrow channel.<ref name="Xiao" /> This channel allows these two components to interact and complete the methyltransfer. The active site in general is considerably tyrosine rich. Residues Tyr245, His297, Ser268, Tyr305, Tyr335, and Tyr337 all help to shape the active site and the channel.<ref name="Xiao" /> The cofactor involved, SAM, provides the methyl for methylation of the lysine on its sulfur atom. |
| + | |||
| + | The beta hairpin stabilizes the conformation of Tyr335 and Tyr337, while also shaping one side of the channel which the peptide binds to.<ref name="Xiao" /> The peptide binding groove is composed of residues 255-268.<ref name="Xiao" /> Lysine would have trouble coming down into the active site in its charged form, but it is facilitated by the faces of the flanking tyrosines.<ref name="Xiao" /> The orientation of the lysine is such that the amine-methyl bond is aligned towards the sulfur on SAM so that it can provide the methyl. There is an important water in the active site as well that acts as a stabilizer for lysine, and helps to shift the lone pair on the nitrogen towards the sulfur of SAM. <ref name="Xiao" /> | ||
| Line 38: | Line 42: | ||
==Student Contributors== | ==Student Contributors== | ||
| - | Ashley Crotteau | + | Ashley Crotteau |
| + | |||
Parker Hiday | Parker Hiday | ||
| + | |||
Lauren Allman | Lauren Allman | ||
Revision as of 17:29, 5 April 2019
H. sapiens Lysine Methyltransferase, SET 7/9
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003 Jun;28(6):329-35. PMID:12826405
- ↑ 2.0 2.1 2.2 Yeates TO. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002 Oct 4;111(1):5-7. PMID:12372294
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 4.0 4.1 Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998 Jun 26;273(26):15933-9. PMID:9632640
- ↑ 5.0 5.1 doi: https://dx.doi.org/10.1016/C2014-0-02189-2
- ↑ https://en.wikipedia.org/wiki/SET_domain#Structure
Student Contributors
Ashley Crotteau
Parker Hiday
Lauren Allman