User:Ashley Crotteau/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
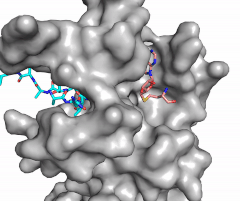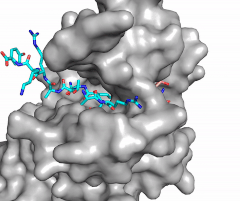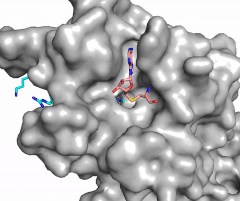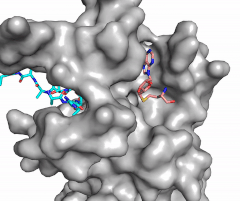
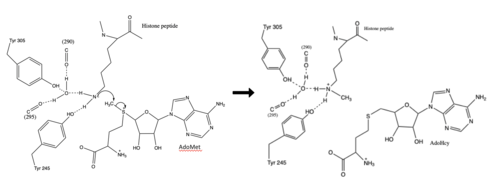
The N of the amine from the lysine serves as a nucleophile that attacks the electrophilic CH<sub>3</sub> that is present in the AdoMet (SAM). The sulfur that the CH<sub>3</sub> is attached to pulls the electrons towards itself to weaken the bond between the sulfur and the carbon. This weak bond allows for the N to break that bond and take the methyl group. The N on the lysine is being stabilized by Tyr residues and a water molecule (Figure 3). This allows the N to accept the methyl and take up that positive charge. | The N of the amine from the lysine serves as a nucleophile that attacks the electrophilic CH<sub>3</sub> that is present in the AdoMet (SAM). The sulfur that the CH<sub>3</sub> is attached to pulls the electrons towards itself to weaken the bond between the sulfur and the carbon. This weak bond allows for the N to break that bond and take the methyl group. The N on the lysine is being stabilized by Tyr residues and a water molecule (Figure 3). This allows the N to accept the methyl and take up that positive charge. | ||
| - | The tyrosine conserved regions of KMT have been found to be vital for only monomethylation. Multiple studies have found that a mutation to any of the conserved tyrosine residues does still monomethylate SAM, but it also creates di-methylation and tri- methylation of SAM.<ref name="Del Rizzo" /> In a recent study, Y245A and Y305F were created through site-directed mutagenesis.<ref name="Del Rizzo" /> Due to size, Ala245 was found to create a larger opening of the channel than tyrosine, which allowed for further methylation of SAM. <ref name="Del Rizzo" /> However, Y305F also showed the same characteristics of di- and tri-methylation, most likely due to a decrease of tyrosine residues shaping the narrow channel.<ref name="Del Rizzo" /> As there are four invariant conserved tyrosine residues (Tyr305, Tyr245, Tyr335, Tyr337) in the active site, this finding indicates that the function of KMT is dependent on the presence of tyrosine residues in the active site. <ref name="Del Rizzo" /> | + | The tyrosine conserved regions of KMT have been found to be vital for only monomethylation. Multiple studies have found that a mutation to any of the conserved tyrosine residues does still monomethylate SAM, but it also creates di-methylation and tri- methylation of SAM.<ref name="Del Rizzo" /> In a recent study, Y245A and Y305F were created through site-directed mutagenesis.<ref name="Del Rizzo" /> Due to size, Ala245 was found to create a larger opening of the channel than tyrosine, which allowed for further methylation of SAM. <ref name="Del Rizzo" /> However, Y305F also showed the same characteristics of di- and tri-methylation, most likely due to a decrease of tyrosine residues shaping the narrow channel.<ref name="Del Rizzo" /> As there are four invariant conserved tyrosine residues (Tyr305, Tyr245, Tyr335, Tyr337) in the active site, this finding indicates that the function of KMT is dependent on the presence of tyrosine residues in the active site. <ref name="Del Rizzo" /> Although only two of the tyrosines are interacting in the mechanism, the other two are shaping the narrow channel so without these extra tyrosines the channel is not as narrow and can therefore di- and tri- methylate lysine. |
== Relevance == | == Relevance == | ||
Revision as of 19:57, 24 April 2019
H. sapiens Lysine Methyltransferase, SET 7/9
| |||||||||||
References
[3] [8] [5] [6] [7] [9] [10] [11] [1] [2] [4]
- ↑ 1.0 1.1 DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 Marino-Ramirez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev Proteomics. 2005 Oct;2(5):719-29. PMID:16209651 doi:http://dx.doi.org/10.1586/14789450.2.5.719
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 4.0 4.1 4.2 doi: https://dx.doi.org/10.15406/mojcsr.2016.03.00047
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003 Jun;28(6):329-35. PMID:12826405
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Yeates TO. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002 Oct 4;111(1):5-7. PMID:12372294
- ↑ 7.0 7.1 Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998 Jun 26;273(26):15933-9. PMID:9632640
- ↑ 8.0 8.1 doi: https://dx.doi.org/10.1016/C2014-0-02189-2
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 Del Rizzo PA, Couture JF, Dirk LM, Strunk BS, Roiko MS, Brunzelle JS, Houtz RL, Trievel RC. SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation. J Biol Chem. 2010 Oct 8;285(41):31849-58. Epub 2010 Aug 1. PMID:20675860 doi:http://dx.doi.org/10.1074/jbc.M110.114587
- ↑ 10.0 10.1 10.2 10.3 10.4 Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010 Dec;21(12):2069-80. doi: 10.1681/ASN.2010060633. Epub 2010, Oct 7. PMID:20930066 doi:http://dx.doi.org/10.1681/ASN.2010060633
- ↑ 11.0 11.1 Tian X, Zhang S, Liu HM, Zhang YB, Blair CA, Mercola D, Sassone-Corsi P, Zi X. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013 Jun;13(5):558-79. doi:, 10.2174/1568009611313050007. PMID:23713993 doi:http://dx.doi.org/10.2174/1568009611313050007
Student Contributors
Ashley Crotteau
Parker Hiday
Lauren Allman