User:Ashley Crotteau/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
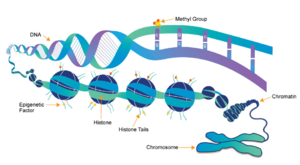
[[Image:Epigentics pic.png|300 px|right|thumb|Figure 1: Epigenetic overview. Showing the methylation of histones and how this affects DNA packing.<ref name="Epigenetics" />]] | [[Image:Epigentics pic.png|300 px|right|thumb|Figure 1: Epigenetic overview. Showing the methylation of histones and how this affects DNA packing.<ref name="Epigenetics" />]] | ||
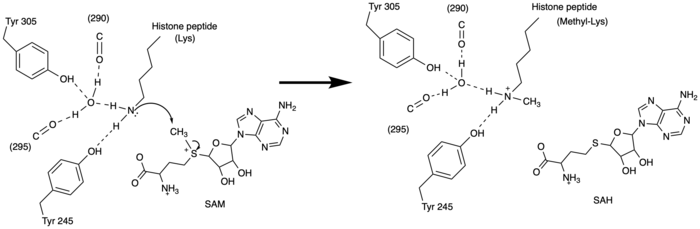
| - | [https://en.wikipedia.org/wiki/Histone Histones] are a family of proteins that condense DNA into chromatin, and is an octamer composed of two of each protein core; H2A, H2B, H3, and H4. Histones are a globular protein, that have N- or C-terminal tails. These tails can be subjected to modifications by enzymes. Methylation of histones is one of the four common histone modifications. Methylation is most common on long tails of H3 and H4 due to the tail being able to enter the active site.<ref name ="DesJarlais" /> A histone can be mono-, di-, or tri- methylated. Once the histone is methylated, the DNA goes from tightly bound heterochromatin to loosely packed euchromatin shown in Figure 1. The euchromatin allows RNA pol II to bind to the DNA and start transcription. <ref name="Marino" /> Histone methylation is | + | [https://en.wikipedia.org/wiki/Histone Histones] are a family of proteins that condense DNA into chromatin, and is an octamer composed of two of each protein core; H2A, H2B, H3, and H4. Histones are a globular protein, that have N- or C-terminal tails. These tails can be subjected to modifications by enzymes. Methylation of histones is one of the four common histone modifications. Methylation is most common on long tails of H3 and H4 due to the tail being able to enter the active site.<ref name ="DesJarlais" /> A histone can be mono-, di-, or tri- methylated. Once the histone is methylated, the DNA goes from tightly bound heterochromatin to loosely packed euchromatin shown in Figure 1. The euchromatin allows RNA pol II to bind to the DNA and start transcription. <ref name="Marino" /> Histone methylation is a major epigenetic marker, which can be passed down from generation to generation. An [https://www.whatisepigenetics.com/fundamentals/ epigenetic marker] affects the way that genes are expressed, and can activate or repress DNA. Histone methylation is a major epigenetic marker because it has the ability to change heterochromatin to euchromatin, and vice versa. Alterations in markers have been associated with many diseases.<ref name="Xiao" /> Lysine Methyltrasferase SET7/9 (KMT) is an enzyme that methylates the histone 3 lysine 4 (H3K4) and plays a important role in the transcription of DNA in ''Homo sapiens''.<ref name="Biterge" /> The methylation of H3K4 results in transcriptional activation.<ref name="Biterge" /> The specific methylation of H3K4 does not result in a change in charge because it is a nonpolar group being added to the lysine. A change in charge could result in tighter bound heterochromatin. |
==Structure== | ==Structure== | ||
| - | The human lysine methyltransferase(HKMT) SET7/9 is 366 amino acids long that comes from ''H. sapiens''. Structure has two proteins in the asymmetric unit, yet acts as a monomer. HKMT will crystalize as a dimer. The structure is composed of the ΔSET7/9 domain. There is a two-domain architecture containing a conserved anti-parallel 𝛃-barrel and an unusual knot-like structure that creates the active site. It also contains a cofactor, SAM, that plays a role in the active site. HKMT only monomethylates H3K4, because the tyrosine rich active site does not allow di- or tri- methylation. | + | The human lysine methyltransferase(HKMT) SET7/9 is 366 amino acids long that comes from ''H. sapiens''. Structure has two proteins in the asymmetric unit, yet acts as a monomer. HKMT will crystalize as a dimer. The structure is composed of the ΔSET7/9 domain. There is a two-domain architecture containing a conserved anti-parallel 𝛃-barrel and an unusual knot-like structure that creates the active site. It also contains a cofactor, SAM, that plays a role in the active site.<ref name="Biterge" /> HKMT only monomethylates H3K4, because the tyrosine rich active site does not allow for di- or tri- methylation. |
===SET Domain=== | ===SET Domain=== | ||
| - | The ΔSET7/9 consists of the SET domain along with the pre- and post-SET regions.<ref name="Schubert" /><ref name="Yeates" /> The pre- and post-SET regions are adjacent to SET domain and are cysteine rich.<ref name="Schubert" /><ref name="Yeates" /> The pre-SET cysteine region is located near the N-terminal where the post-SET region is located near the C-terminal of the domain.<ref name="Schubert" /><ref name="Yeates" /> These regions are said to play an important role in substrate recognition and enzymatic activity.<ref name="Schubert" /><ref name="Yeates" /> The cysteine regions are not shown in the crystal structure as only residues 117-366 were crystalized.<ref name="Xiao" /> | + | The ΔSET7/9 consists of the [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1273623/ SET domain] along with the pre- and post-SET regions.<ref name="Schubert" /><ref name="Yeates" /> The pre- and post-SET regions are adjacent to SET domain and are cysteine rich.<ref name="Schubert" /><ref name="Yeates" /> The pre-SET cysteine region is located near the N-terminal where the post-SET region is located near the C-terminal of the domain.<ref name="Schubert" /><ref name="Yeates" /> These regions are said to play an important role in substrate recognition and enzymatic activity.<ref name="Schubert" /><ref name="Yeates" /> The cysteine regions are not shown in the crystal structure as only residues 117-366 were crystalized.<ref name="Xiao" /> |
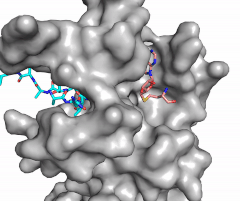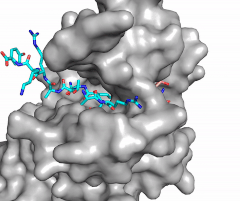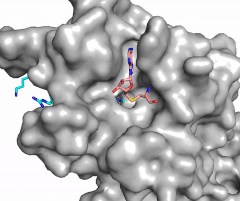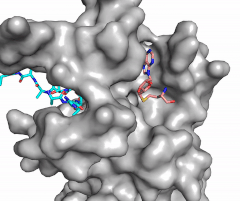
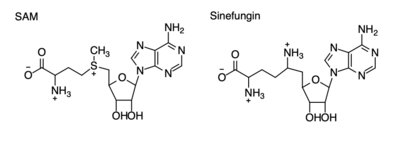
The SET domain is mostly defined by turns and loops, which connect secondary structures, with the few <scene name='81/811707/Beta_sheets/4'>antiparallel β-sheets</scene>.<ref name="Schubert" /> <scene name='81/811707/Beta-hairpin/3'>Residues 337-349</scene> form a β-hairpin that sticks out at a right angle to the surface of the enzyme.<ref name="Xiao" /> This hairpin stabilizes the conformation of tyrosine residues.<ref name="Xiao" /> The following three residues (<scene name='81/811707/Sharp_bend/3'>350-352</scene>) accommodate a unique sharp bend in the peptide chain and the end of the protein takes on an <scene name='81/811707/C-term_alpha_helix/2'>α-helical conformation</scene>.<ref name="Xiao" /> This alpha helix packs against a beta sheet and orients the cofactor towards the active site.<ref name="Xiao" /> The two most defining features of the SET domain are the C-terminal tyrosine and the <scene name='81/811707/Variable_knot/3'>knot-like fold</scene>. These two components have been recognized to be essential for S-adenosyl-L-methionine(SAM) binding and catalysis, which is shown as <scene name='81/811707/Sam_isolated/3'>S-adenosyl-L-homocysteine</scene>(SAH) in the structure after methylation of the histone.<ref name="Schubert" /> <ref name="Yeates" /> <ref name="Huang" /> The knot-like fold contains the binding sites for the cofactor SAM and the peptide substrate.<ref name="Licciardello" /> | The SET domain is mostly defined by turns and loops, which connect secondary structures, with the few <scene name='81/811707/Beta_sheets/4'>antiparallel β-sheets</scene>.<ref name="Schubert" /> <scene name='81/811707/Beta-hairpin/3'>Residues 337-349</scene> form a β-hairpin that sticks out at a right angle to the surface of the enzyme.<ref name="Xiao" /> This hairpin stabilizes the conformation of tyrosine residues.<ref name="Xiao" /> The following three residues (<scene name='81/811707/Sharp_bend/3'>350-352</scene>) accommodate a unique sharp bend in the peptide chain and the end of the protein takes on an <scene name='81/811707/C-term_alpha_helix/2'>α-helical conformation</scene>.<ref name="Xiao" /> This alpha helix packs against a beta sheet and orients the cofactor towards the active site.<ref name="Xiao" /> The two most defining features of the SET domain are the C-terminal tyrosine and the <scene name='81/811707/Variable_knot/3'>knot-like fold</scene>. These two components have been recognized to be essential for S-adenosyl-L-methionine(SAM) binding and catalysis, which is shown as <scene name='81/811707/Sam_isolated/3'>S-adenosyl-L-homocysteine</scene>(SAH) in the structure after methylation of the histone.<ref name="Schubert" /> <ref name="Yeates" /> <ref name="Huang" /> The knot-like fold contains the binding sites for the cofactor SAM and the peptide substrate.<ref name="Licciardello" /> | ||
Revision as of 23:02, 25 April 2019
H. sapiens Lysine Methyltransferase, SET 7/9
| |||||||||||
References
[4] [9] [6] [7] [8] [10] [11] [12] [2] [3] [5] [1]
- ↑ 1.0 1.1 https://www.whatisepigenetics.com/fundamentals/
- ↑ 2.0 2.1 DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 Marino-Ramirez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev Proteomics. 2005 Oct;2(5):719-29. PMID:16209651 doi:http://dx.doi.org/10.1586/14789450.2.5.719
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ 5.0 5.1 5.2 5.3 doi: https://dx.doi.org/10.15406/mojcsr.2016.03.00047
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003 Jun;28(6):329-35. PMID:12826405
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Yeates TO. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002 Oct 4;111(1):5-7. PMID:12372294
- ↑ 8.0 8.1 Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998 Jun 26;273(26):15933-9. PMID:9632640
- ↑ 9.0 9.1 doi: https://dx.doi.org/10.1016/C2014-0-02189-2
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 Del Rizzo PA, Couture JF, Dirk LM, Strunk BS, Roiko MS, Brunzelle JS, Houtz RL, Trievel RC. SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation. J Biol Chem. 2010 Oct 8;285(41):31849-58. Epub 2010 Aug 1. PMID:20675860 doi:http://dx.doi.org/10.1074/jbc.M110.114587
- ↑ 11.0 11.1 11.2 11.3 11.4 Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol. 2010 Dec;21(12):2069-80. doi: 10.1681/ASN.2010060633. Epub 2010, Oct 7. PMID:20930066 doi:http://dx.doi.org/10.1681/ASN.2010060633
- ↑ 12.0 12.1 Tian X, Zhang S, Liu HM, Zhang YB, Blair CA, Mercola D, Sassone-Corsi P, Zi X. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: new targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013 Jun;13(5):558-79. doi:, 10.2174/1568009611313050007. PMID:23713993 doi:http://dx.doi.org/10.2174/1568009611313050007
Student Contributors
Ashley Crotteau
Parker Hiday
Lauren Allman