This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Proteins from Mycobacterium tuberculosis
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='' size='450' side='right' scene='80/801748/Cv/1' caption=''> | <StructureSection load='' size='450' side='right' scene='80/801748/Cv/1' caption=''> | ||
| + | ===Novel T9 loop interaction of Filamenting Temperature-sensitive mutant Z from ''Mycobacterium tuberculosis''<ref>doi 10.1107/S2053230X19004618</ref>=== | ||
| + | |||
| + | As of 2017, tuberculosis has infected 1.7 billion people (23% of the world's population) and has caused 10 million deaths. ''Mycobacterium tuberculosis'' (Mtb) is quickly evolving, and new strains are classified as multi-drug resistant. Thus, the development and discovery of new drugs to combat Mtb is vital to combat the drug-resistant strains. Filamenting temperature-sensitive mutant Z (FtsZ), an important protein involved in cell-division is key for the survival of Mtb. Here, we have solved the crystal structure of MtbFtsZ that exhibit an inter-subunit that plays a biological role in the GTPase activity of MtbFtsZ and have elucidated a novel conformation, involving the T9 loop and the nucleotide binding pocket that breaks up the GTPase active site. This novel conformation can serve as basis for the development of the novel drugs to combat tuberculosis. | ||
| + | |||
| + | <scene name='81/813404/Cv/2'>Asymmetric unit contains 6 protomers that form a dimer of trimers</scene> ([[5v68]]). Protomers A and C shown in blue, protomer B shown in gray, protomers D and F shown in red, protomer E shown in cyan, GDP are represented by orange spheres, and PO4 by green spheres. <scene name='81/813404/Cv/4'>Trimer ABC superimposed onto DEF</scene>. Same color scheme as in previous scene. | ||
| + | |||
| + | Comparison of other MtbFtsZ structures with trimer ABC from [[5v68]] reveals that protomers A and B superimposed well but the conformation of protomer C is vastly different. <scene name='81/813404/Cv/23'>Structure 5V68 superimposed onto 4KWE</scene>. Nucleotide for both is GDP. Color scheme is as follows, [[5v68]] blue, [[4kwe]] yellow, nucleotides orange spheres, Glu231 green spheres, phosphate red spheres. <scene name='81/813404/Cv/24'>Click here to see animation of this scene</scene>. <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Toggle Animation</text> | ||
| + | </jmolButton></jmol> The trimer ABC from [[5v68]] is to a curved MtbFtsZ protofilament (PDB [[4kwe]]). Based on an r.m.s.d. of 1.2Å between protomers AB of [[5v68]] and protomers CB of [[4kwe]], protomers A and B from [[5v68]] are similar with protomers C and B of [[4kwe]]. <scene name='81/813404/Cv/25'>Glu231 (shown as green sphere) from protomer C of [[5v68]] and Glu231 from protomer A of [[4kwe]] are far apart</scene>. | ||
| + | |||
| + | <scene name='81/813404/Cv/26'>5V68 with bound GDP superimposed onto 2Q1Y with bound GTP</scene>. Color scheme is as follows, [[5v68]] blue, [[2q1y]] green, nucleotides orange spheres, Glu231 green spheres, phosphate red spheres. <scene name='81/813404/Cv/27'>Click here to see animation of this scene</scene>. <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Toggle Animation</text> | ||
| + | </jmolButton></jmol> To generate this trimer ([[2q1y]]), A1 A2 A3, the crystal symmetry molecules neighboring chain A2 that exhibit the inter-subunit interface were used. Only chains A1 and A2 were used for the superimposition. Again, protomers A and B from present study are similar with the top two protomers A1 and A2 from [[2q1y]] with an r.m.s.d of 2.1Å. <scene name='81/813404/Cv/29'>The distance of their respective Glu231</scene> is approximately 63Å. | ||
| + | |||
| + | <scene name='81/813404/Cv/21'>4KWE with bound GDP superimposed onto 2Q1Y with bound GTP</scene>. Color scheme is as follows, [[4kwe]] yellow, [[2q1y]] green, nucleotides orange spheres, Glu231 green spheres. <scene name='81/813404/Cv/22'>Click here to see animation of this scene</scene>. <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Toggle Animation</text> | ||
| + | </jmolButton></jmol> Superimposition of [[4kwe]] and [[2q1y]] reveals an r.m.s.d. of 2.8Å between the trimers and a 12.5Å <scene name='81/813404/Cv/30'>distance between their respective Glu231’s of A and A3</scene>, showing that these structures are relatively similar. The distances between Glu231 of [[5v68]] from protomer C is far greater than 12.5Å and the interaction with the middle protomers of [[4kwe]] and [[2q1y]] with protomer C of [[5v68]] involve the T9 loop residue Glu231, which is not the case in the other structures. | ||
| + | |||
| + | <scene name='81/813404/Cv1/3'>Crystal structure of protomers A and B of 5V68</scene> (gray). <scene name='81/813404/Cv1/6'>A zoom-in of the inter-subunit interface with bound GDP</scene> (orange). Helix H11 is shown in green. Helices η1, H7, and loop T6 are shown in blue. Switch I (T3 loop) is disordered surrounded by a cloud. Switch II (sH2) is shown in magenta. The top two protomers of all three structures (AB for [[5v68]], A1 A2 for [[2q1y]], and CB for [[4kwe]]) all exhibit a similar inter-subunit interface. This interaction between protomers A and B of 5V68 involve the T6 and T7 loops, helices: H11, η1, and H7. Protomer A of our structure “sits” on the helices η1, H7 and loop T6 (all shown in blue) from protomer B. This brings the T7 loop’s (shown in red) residues <scene name='81/813404/Cv1/9'>Asn205, Asp207 and Asp210</scene> of protomer A of [[5v68]] within 16Å of GDP from protomer B, forming the GTPase active site. The T3 loop is disordered or in its OFF position (surrounded by magenta cloud). Helix sH2 is also OFF because there is no hydrogen network (shown in magenta). | ||
| + | |||
| + | <scene name='81/813404/Cv1/11'>Crystal structure of protomer B and C of 5V68</scene>. Protomer B is shown in gray, the T9 loop in brown, T11 in cyan; and the N-terminal of protomer C is shown in green, C-terminal blue, helix H8 yellow, the T7 loop red, and the switches in magenta. <scene name='81/813404/Cv1/12'>PO4 binding site</scene>. Glu231 from protomer B is shown in brown, while Glu274 from protomer is in gray. The residues in green are from protomer C. | ||
| + | |||
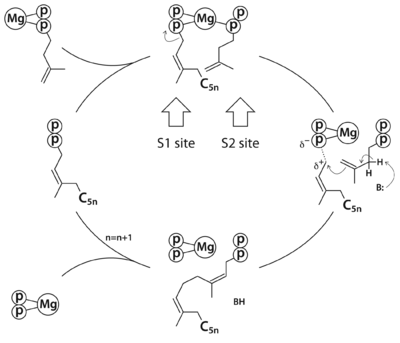
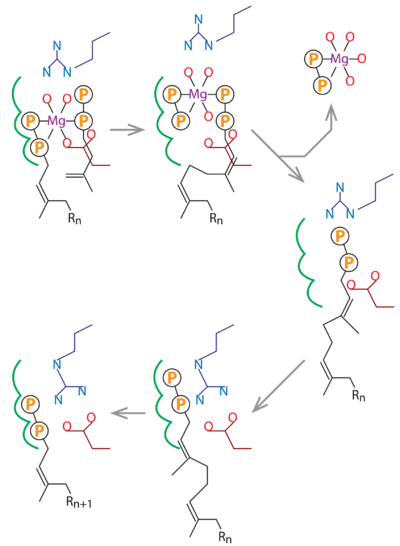
===Substrate analogue complex structure of ''Mycobacterium tuberculosis'' decaprenyl diphosphate synthase<ref>doi 10.1107/S2053230X19001213</ref> === | ===Substrate analogue complex structure of ''Mycobacterium tuberculosis'' decaprenyl diphosphate synthase<ref>doi 10.1107/S2053230X19001213</ref> === | ||
Revision as of 12:37, 28 May 2019
| |||||||||||
References
- ↑ Lazo EO, Jakoncic J, RoyChowdhury S, Awasthi D, Ojima I. Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis. Acta Crystallogr F Struct Biol Commun. 2019 May 1;75(Pt 5):359-367. doi:, 10.1107/S2053230X19004618. Epub 2019 Apr 24. PMID:31045565 doi:http://dx.doi.org/10.1107/S2053230X19004618
- ↑ Ko TP, Xiao X, Guo RT, Huang JW, Liu W, Chen CC. Substrate-analogue complex structure of Mycobacterium tuberculosis decaprenyl diphosphate synthase. Acta Crystallogr F Struct Biol Commun. 2019 Apr 1;75(Pt 4):212-216. PMID:30950820 doi:10.1107/S2053230X19001213
- ↑ Gupta AK, Behera D, Gopal B. The crystal structure of Mycobacterium tuberculosis high-temperature requirement A protein reveals an autoregulatory mechanism. Acta Crystallogr F Struct Biol Commun. 2018 Dec 1;74(Pt 12):803-809. doi:, 10.1107/S2053230X18016217. Epub 2018 Nov 29. PMID:30511675 doi:http://dx.doi.org/10.1107/S2053230X18016217
- ↑ Hasenbein S, Meltzer M, Hauske P, Kaiser M, Huber R, Clausen T, Ehrmann M. Conversion of a regulatory into a degradative protease. J Mol Biol. 2010 Apr 9;397(4):957-66. doi: 10.1016/j.jmb.2010.02.027. Epub 2010, Feb 22. PMID:20184896 doi:http://dx.doi.org/10.1016/j.jmb.2010.02.027
- ↑ Sohn J, Grant RA, Sauer RT. OMP peptides activate the DegS stress-sensor protease by a relief of inhibition mechanism. Structure. 2009 Oct 14;17(10):1411-21. PMID:19836340 doi:10.1016/j.str.2009.07.017
- ↑ Ash EL, Sudmeier JL, Day RM, Vincent M, Torchilin EV, Haddad KC, Bradshaw EM, Sanford DG, Bachovchin WW. Unusual 1H NMR chemical shifts support (His) C(epsilon) 1...O==C H-bond: proposal for reaction-driven ring flip mechanism in serine protease catalysis. Proc Natl Acad Sci U S A. 2000 Sep 12;97(19):10371-6. PMID:10984533
- ↑ Radisky ES, Lee JM, Lu CJ, Koshland DE Jr. Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6835-40. Epub 2006 Apr 24. PMID:16636277
- ↑ Dym O, Albeck S, Peleg Y, Schwarz A, Shakked Z, Burstein Y, Zimhony O. Structure-function analysis of the acyl carrier protein synthase (AcpS) from Mycobacterium tuberculosis. J Mol Biol. 2009 Nov 6;393(4):937-50. Epub 2009 Sep 3. PMID:19733180 doi:10.1016/j.jmb.2009.08.065