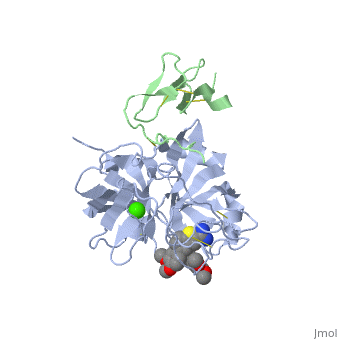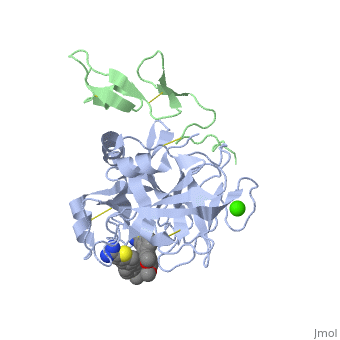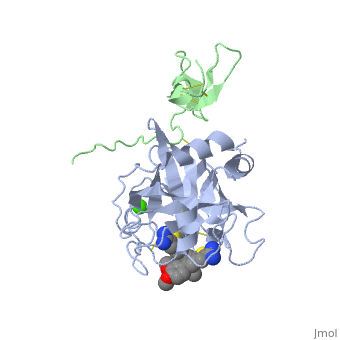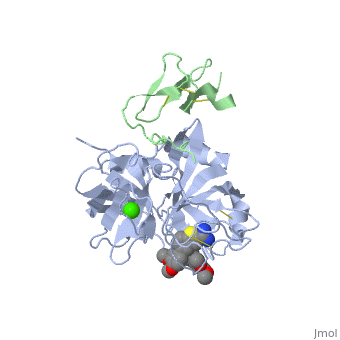
Factor IX
From Proteopedia
(Difference between revisions)
| Line 154: | Line 154: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of factor IX== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Factor IX; Domains – GLA 1-47; EGF-like 92-130; heavy 227-461; light 133-185 | ||
| - | |||
| - | **[[1mgx]], [[1cfi]], [[1cfh]] - hIX GLA domain – human – NMR<br /> | ||
| - | **[[1edm]], [[5vyg]] – hIX EGF-like domain<br /> | ||
| - | **[[1ixa]] - hIX EGF-like domain - NMR<br /> | ||
| - | **[[6mv4]] - hIX light+heavy chain<br /> | ||
| - | |||
| - | *Factor IX complexes | ||
| - | |||
| - | **[[3lc3]], [[3lc5]] – hIXa light+heavy chain + benzothiophene inhibitor<br /> | ||
| - | **[[5vgy]] - hIXa EGF-like domain + trisaccharide<br /> | ||
| - | **[[3kcg]] - hIXa light+heavy chains + antithrombin-III+ pentasaccharide<br /> | ||
| - | **[[2wph]], [[2wpi]], [[2wpj]], [[2wpk]], [[2wpl]] - hIXa light+heavy chains (mutant) + FPR-chloromethyl ketone<br /> | ||
| - | **[[2wpm]] - hIXa light+heavy chains (mutant) + EGR-chloromethyl ketone<br /> | ||
| - | **[[1nl0]] – hIX GLA domain + anti-factor IX antibody<br /> | ||
| - | **[[1rfn]] - hIXa light+heavy chains EGF2-catalytic domain + inhibitor<br /> | ||
| - | **[[4yzu]], [[4z0k]], [[5tno]] – hIX peptidase S1 and EGF-like 2 domains + inhibitor <br /> | ||
| - | **[[4zae]], [[5egm]], [[5jb8]], [[5jb9]], [[5jba]], [[5jbb]], [[5jbc]], [[5tnt]] – hIX peptidase S1 and EGF-like 2 domains (mutant) + inhibitor <br /> | ||
| - | **[[4wm0]] – hIX EGF-like domain + xyloside gxylosyltransferase + glucopyranose derivative<br /> | ||
| - | **[[4wma]] – hIX EGF-like domain + xyloside gxylosyltransferase + UDP-glucose <br /> | ||
| - | **[[4wnh]] – hIX EGF-like domain + xyloside gxylosyltransferase + UDP-xylose <br /> | ||
| - | **[[4wmb]] – hIX EGF-like domain + xyloside gxylosyltransferase + UDP <br /> | ||
| - | **[[4wmi]], [[4wmk]], [[4wn2]] – hIX EGF-like domain + xyloside gxylosyltransferase + glucopyranose derivative + UDP<br /> | ||
| - | **[[5f86]] – hIX EGF-like domain + glucosyltransferase <br /> | ||
| - | **[[5f84]] – hIX EGF-like domain + glucosyltransferase + UDP-glucose <br /> | ||
| - | **[[5f85]] – hIX EGFlike domain + glucosyltransferase + UDP <br /> | ||
| - | **[[6rfk]] – hIX light+heavy chains + peptide - NMR <br /> | ||
| - | **[[1x7a]] - pIXa light+heavy chains + inhibitor – pig<br /> | ||
| - | **[[1pfx]] – pIXa +FPR<br /> | ||
| - | **[[1j34]], [[1j35]] - bIX GLA domain + IX-binding protein – bovine<br /> | ||
| - | **[[1pfx]] – pIXa +FPR<br /> | ||
| - | }} | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 06:49, 26 June 2019
| |||||||||||
Additional Resources
For additional information, see: Colored & Bioluminescent Proteins
For additional information, see: Hemophilia
References
- ↑ Spitzer SG, Kuppuswamy MN, Saini R, Kasper CK, Birktoft JJ, Bajaj SP. Factor IXHollywood: substitution of Pro55 by Ala in the first epidermal growth factor-like domain. Blood. 1990 Oct 15;76(8):1530-7. PMID:2169923
- ↑ Schmidt AE, Bajaj SP. Structure-function relationships in factor IX and factor IXa. Trends Cardiovasc Med. 2003 Jan;13(1):39-45. PMID:12554099
- ↑ Zhong D, Bajaj MS, Schmidt AE, Bajaj SP. The N-terminal epidermal growth factor-like domain in factor IX and factor X represents an important recognition motif for binding to tissue factor. J Biol Chem. 2002 Feb 1;277(5):3622-31. Epub 2001 Nov 26. PMID:11723140 doi:10.1074/jbc.M111202200
- ↑ Perona JJ, Craik CS. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J Biol Chem. 1997 Nov 28;272(48):29987-90. PMID:9374470
- ↑ Ludwig M, Sabharwal AK, Brackmann HH, Olek K, Smith KJ, Birktoft JJ, Bajaj SP. Hemophilia B caused by five different nondeletion mutations in the protease domain of factor IX. Blood. 1992 Mar 1;79(5):1225-32. PMID:1346975
- ↑ Murav'ev IA, Mar'iasis ED, Starokozhko LE, Chebotarev VV, Krasova TG. [Determination of the biologic availability of preparations for topical use by experimental dermatologic methods] Farmatsiia. 1977 Jul-Aug;26(4):15-9. PMID:902786
- ↑ Morris DP, Stevens RD, Wright DJ, Stafford DW. Processive post-translational modification. Vitamin K-dependent carboxylation of a peptide substrate. J Biol Chem. 1995 Dec 22;270(51):30491-8. PMID:8530480
- ↑ Pan LC, Price PA. The propeptide of rat bone gamma-carboxyglutamic acid protein shares homology with other vitamin K-dependent protein precursors. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6109-13. PMID:3875856
- ↑ Knobloch JE, Suttie JW. Vitamin K-dependent carboxylase. Control of enzyme activity by the "propeptide" region of factor X. J Biol Chem. 1987 Nov 15;262(32):15334-7. PMID:2890628
- ↑ Knobloch JE, Suttie JW. Vitamin K-dependent carboxylase. Control of enzyme activity by the "propeptide" region of factor X. J Biol Chem. 1987 Nov 15;262(32):15334-7. PMID:2890628
- ↑ Cheung A, Engelke JA, Sanders C, Suttie JW. Vitamin K-dependent carboxylase: influence of the "propeptide" region on enzyme activity. Arch Biochem Biophys. 1989 Nov 1;274(2):574-81. PMID:2802629
- ↑ Lin PJ, Jin DY, Tie JK, Presnell SR, Straight DL, Stafford DW. The putative vitamin K-dependent gamma-glutamyl carboxylase internal propeptide appears to be the propeptide binding site. J Biol Chem. 2002 Aug 9;277(32):28584-91. Epub 2002 May 28. PMID:12034728 doi:10.1074/jbc.M202292200
- ↑ Sanford DG, Kanagy C, Sudmeier JL, Furie BC, Furie B, Bachovchin WW. Structure of the propeptide of prothrombin containing the gamma-carboxylation recognition site determined by two-dimensional NMR spectroscopy. Biochemistry. 1991 Oct 15;30(41):9835-41. PMID:1911775
- ↑ Lin PJ, Jin DY, Tie JK, Presnell SR, Straight DL, Stafford DW. The putative vitamin K-dependent gamma-glutamyl carboxylase internal propeptide appears to be the propeptide binding site. J Biol Chem. 2002 Aug 9;277(32):28584-91. Epub 2002 May 28. PMID:12034728 doi:10.1074/jbc.M202292200
- ↑ Furie B, Bouchard BA, Furie BC. Vitamin K-dependent biosynthesis of gamma-carboxyglutamic acid. Blood. 1999 Mar 15;93(6):1798-808. PMID:10068650
- ↑ Thompson AR. Structure, function, and molecular defects of factor IX. Blood. 1986 Mar;67(3):565-72. PMID:3511981
- ↑ Stenflo J, Fernlund P, Egan W, Roepstorff P. Vitamin K dependent modifications of glutamic acid residues in prothrombin. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2730-3. PMID:4528109
- ↑ Furie B, Furie BC. Molecular and cellular biology of blood coagulation. N Engl J Med. 1992 Mar 19;326(12):800-6. PMID:1538724 doi:http://dx.doi.org/10.1056/NEJM199203193261205
- ↑ Shikamoto Y, Morita T, Fujimoto Z, Mizuno H. Crystal structure of Mg2+- and Ca2+-bound Gla domain of factor IX complexed with binding protein. J Biol Chem. 2003 Jun 27;278(26):24090-4. Epub 2003 Apr 14. PMID:12695512 doi:10.1074/jbc.M300650200
- ↑ Huang M, Furie BC, Furie B. Crystal structure of the calcium-stabilized human factor IX Gla domain bound to a conformation-specific anti-factor IX antibody. J Biol Chem. 2004 Apr 2;279(14):14338-46. Epub 2004 Jan 13. PMID:14722079 doi:10.1074/jbc.M314011200
- ↑ Golinelli-Pimpaneau B, Gigant B, Bizebard T, Navaza J, Saludjian P, Zemel R, Tawfik DS, Eshhar Z, Green BS, Knossow M. Crystal structure of a catalytic antibody Fab with esterase-like activity. Structure. 1994 Mar 15;2(3):175-83. PMID:8069632
- ↑ Furie BC, Blumenstein M, Furie B. Metal binding sites of a gamma-carboxyglutamic acid-rich fragment of bovine prothrombin. J Biol Chem. 1979 Dec 25;254(24):12521-30. PMID:500729
- ↑ Nakagawa T, Tani M, Kita K, Ito M. Preparation of fluorescence-labeled GM1 and sphingomyelin by the reverse hydrolysis reaction of sphingolipid ceramide N-deacylase as substrates for assay of sphingolipid-degrading enzymes and for detection of sphingolipid-binding proteins. J Biochem. 1999 Sep;126(3):604-11. PMID:10467178
Proteopedia Page Contributors and Editors (what is this?)
Nadia Dorochko, Michal Harel, Alexander Berchansky, David Canner