Helicase
From Proteopedia
| Line 62: | Line 62: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of helicase== | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Helicase Mot1 | ||
| - | |||
| - | **[[3oc3]] – Hel MOT1 heat domain + TFIID-1 – ''Encephalitozoon cuniculi''<BR /> | ||
| - | |||
| - | *Helicase 308 | ||
| - | |||
| - | **[[2p6r]] – AfHel 308 + DNA – ''Archaeoglobus fulgidus''<BR /> | ||
| - | **[[2p6u]] - AfHel 308 | ||
| - | |||
| - | *DNA helicase | ||
| - | |||
| - | **[[3fld]] – EcHel I C terminal – ''Escherichia coli''<BR /> | ||
| - | **[[4l0j]] – EcHel I translocation domain<br /> | ||
| - | **[[2q7t]], [[2q7u]] – EcHel I relaxase domain (mutant) <BR /> | ||
| - | **[[1p4d]] - EcHel I relaxase domain<BR /> | ||
| - | **[[4n8o]], [[5n8o]] - EcHel I + DNA<br /> | ||
| - | **[[2a0i]] - EcHel I (mutant) + DNA<BR /> | ||
| - | **[[1e0j]], [[1e0k]] – Hel 4D domain – Phage T7 | ||
| - | |||
| - | *DNA helicase II or UvrD | ||
| - | |||
| - | **[[1q2z]], [[1rw2]] – hHel II KU86 C terminal – human - NMR<BR /> | ||
| - | **[[5y58]] – yHel II + DNA – yeast<br /> | ||
| - | **[[5y59]] – yHel II subunit 2 + Sir4P peptide<br /> | ||
| - | **[[3lfu]] – EcHel II <BR /> | ||
| - | **[[2is1]], [[2is2]] - EcHel II (mutant) + DNA<BR /> | ||
| - | **[[2is4]], [[2is6]] - EcHel II (mutant) + ADP + DNA<BR /> | ||
| - | **[[4c2t]], [[4c2u]], [[4c30]] – DrHel II + DNA - ''Deinococcus radiodurans''<br /> | ||
| - | |||
| - | *ATP-dependent helicase ATRX | ||
| - | |||
| - | **[[3qln]] - hATRX ADD domain residues 167-289 – human<BR /> | ||
| - | **[[2ld1]], [[2jm1]] - hATRX ADD domain - NMR<BR /> | ||
| - | **[[3ql9]], [[4w5a]] – hATRX ADD domain + histone H3 peptide <BR /> | ||
| - | **[[2lbm]] - hATRX ADD domain + histone H3 peptide – NMR<BR /> | ||
| - | **[[3qla]], [[3qlc]] - hATRX ADD domain (mutant) + histone H3 peptide<BR /> | ||
| - | **[[5y6o]] – hATRX N terminal + ATRX<br /> | ||
| - | **[[2xzp]] - hRENT1 helicase domain<BR /> | ||
| - | **[[2wjv]] - hRENT1 helicase CH and C terminal domains<BR /> | ||
| - | **[[2wjy]] - hRENT1 helicase CH domains<BR /> | ||
| - | **[[2xzo]] – hRENT1 helicase domain + RNA<BR /> | ||
| - | |||
| - | *ATP-dependent helicase Fun30 | ||
| - | |||
| - | **[[5gn1]] – yFun C-terminal<br /> | ||
| - | |||
| - | *ATP-dependent helicase Rho | ||
| - | |||
| - | **[[3l0o]] – TmRho – ''Thermotoga maritima''<BR /> | ||
| - | **[[3ice]] – EcRho + ADP + BeF3 + RNA<BR /> | ||
| - | **[[2ht1]] - EcRho + RNA<BR /> | ||
| - | **[[1a8v]], [[1a62]] – EcRho RNA-binding domain<br /> | ||
| - | **[[1a63]] - EcRho RNA-binding domain – NMR<br /> | ||
| - | **[[2a8v]] – EcRho RNA-binding domain + RNA<br /> | ||
| - | **[[1pv4]] – EcRho + DNA<br /> | ||
| - | **[[1pvo]] – EcRho + ssRNA + ANPPNP<br /> | ||
| - | **[[1xpr]], [[1xpu]], [[1xpo]] - EcRho + ssRNA + antibiotic | ||
| - | |||
| - | *ATP-dependent helicase MFD | ||
| - | |||
| - | **[[3hjh]] – EcMFD motor domain | ||
| - | |||
| - | *ATP-dependent helicase HEPA | ||
| - | |||
| - | **[[6bog]] – EcHEPA | ||
| - | |||
| - | *ATP-dependent helicase E1 | ||
| - | |||
| - | **[[2v9p]] – PvE1 helicase domain – Bovine papillomavirus<br /> | ||
| - | **[[2gxa]] – PvE1 + DNA + Mg-ADP<br /> | ||
| - | **[[1tue]] – PvE1 + E2 – human papillomavirus | ||
| - | |||
| - | *ATP-dependent helicase CHD | ||
| - | |||
| - | **[[2b2y]] – hCHD1 <br /> | ||
| - | **[[4b4c]] – hCHD1 DNA-binding domain<br /> | ||
| - | **[[2b2t]], [[2b2u]], [[2b2v]], [[2b2w]] – hCHD1 (mutant) + histone H3 tail<br /> | ||
| - | **[[4nw2]] – hCHD1 + NS1 peptide<br /> | ||
| - | **[[2ee1]] – hCHD4 chromodomain – NMR<br /> | ||
| - | **[[2l5u]] – hCHD4 PHD1 domain – NMR<br /> | ||
| - | **[[2l75]] – hCHD4 PHD2 domain + histone H3 peptide– NMR<br /> | ||
| - | **[[2epb]] – hCHD6 chromodomain – NMR<br /> | ||
| - | **[[2ckc]], [[2v0e]], [[2v0f]] – hCHD7 BRK domain - NMR <br /> | ||
| - | **[[2dl6]] – hCHD8 BRK domain - NMR <br /> | ||
| - | **[[2cka]] – hCHD8 <br /> | ||
| - | **[[2h1e]], [[2xb0]], [[3mwy]] – yCHD1 chromodomain<BR /> | ||
| - | **[[2dy7]] - yCHD1 chromodomain 1 – NMR<BR /> | ||
| - | **[[2dy8]] - yCHD1 chromodomain 2 – NMR<BR /> | ||
| - | **[[3ted]] - yCHD1 chromodomain 1 + DNA<br /> | ||
| - | |||
| - | *ATP-dependent DNA helicase PIF1 | ||
| - | |||
| - | **[[5fhh]], [[6hpt]] – hPIF residues 200-641<br /> | ||
| - | **[[6hpu]], [[6hpq]], [[6hph]] – hPIF residues 206-620 + nucleotide<br /> | ||
| - | **[[5o6b]], [[5o6d]] – yPIF residues 237-780 + DNA + ATP<br /> | ||
| - | **[[5o6e]] – yPIF residues 237-780 + DNA + ADP<br /> | ||
| - | |||
| - | *ATP-dependent DNA helicase TA0057 | ||
| - | |||
| - | **[[5h8w]] – TaTA0 + DNA – ''Thermoplasma acidophilum''<br /> | ||
| - | |||
| - | *ATP-dependent DNA helicase LHR | ||
| - | |||
| - | **[[5v9x]] – MsLHR + DNA – ''Mycobacterium smegmatis''<br /> | ||
| - | |||
| - | *Dead box ATP-dependent RNA helicase | ||
| - | |||
| - | **[[4xw3]] – hDDX1 SPRY domain<br /> | ||
| - | **[[2i4i]] – hDDX3X residues 167-581<BR /> | ||
| - | **[[4px9]] - hDDX3X residues 135-407<br /> | ||
| - | **[[4pxa]] - hDDX3X residues 135-407 (mutant)<br /> | ||
| - | **[[5e7i]] - hDDX3X residues 133-584<br /> | ||
| - | **[[2jgn]] – hDDX3X helicase domain residues 408-579<BR /> | ||
| - | **[[3ly5]] – hDDX18 dead domain<BR /> | ||
| - | **[[2rb4]] – hDDX25 helicase domain<BR /> | ||
| - | **[[2p6n]] – hDDX41 helicase domain (mutant) <BR /> | ||
| - | **[[2e29]], [[2m3d]] - hDDX50 GUCT domain - NMR<BR /> | ||
| - | **[[1vec]] – hP54 N terminal<BR /> | ||
| - | **[[3fho]] – DBP5 – fission yeast<BR /> | ||
| - | **[[2kbe]] – yDBP5 N terminal – NMR<BR /> | ||
| - | **[[2kbf]] - yDBP5 C terminal – NMR<BR /> | ||
| - | **[[3gfp]] - yDBP5 C-terminal<BR /> | ||
| - | **[[2hjv]] – BsDBPA domain 2 – ''Bacillus subtilis''<BR /> | ||
| - | **[[2g0c]] - BsDBPA RNA-binding domain<br /> | ||
| - | **[[3eaq]], [[3ear]], [[3eas]] – TtHera residues 215-426 - ''Thermus thermophilus''<br /> | ||
| - | **[[3i31]] - TtHera residues 431-517<br /> | ||
| - | **[[3i32]] - TtHera residues 218-517<br /> | ||
| - | **[[3nej]] - TtHera N terminal (mutant)<br /> | ||
| - | **[[5ivl]] – GsDB CSHA - ''Geobacillus stearothermophilus''<br /> | ||
| - | **[[5b88]] - EcDBP RRM-like domain - NMR<br /> | ||
| - | **[[5gju]] - EcDBP residues 6-210<br /> | ||
| - | **[[5gi4]] - EcDBP residues 218-445<br /> | ||
| - | **[[6aib]] – SaDB CSHA N-terminal – ''Staphylococcus aureus''<br /> | ||
| - | |||
| - | *''Dead box ATP-dependent RNA helicase complexes'' | ||
| - | |||
| - | **[[3fmo]], [[3fhc]] – hDBP5 + Nup214<BR /> | ||
| - | **[[3fht]] – hDBP5 + AMPPNP + RNA<BR /> | ||
| - | **[[6cz5]] - hDDX3X residues 132-607 + AMP<br /> | ||
| - | **[[6b4k]] – hDDX19B residues 54-479 + ANP<br /> | ||
| - | **[[6b4j]], [[6b4i]] – hDDX19B residues 54-479 + GLE1 + Nup42 + ANP<br /> | ||
| - | **[[3fmp]] – hDDX19B + Nup214<br /> | ||
| - | **[[3g0h]] – hDDX19B residues 54-275 + ATP analog + RNA<BR /> | ||
| - | **[[3ews]] - hDDX19B residues 54-275 + ADP<BR /> | ||
| - | **[[5e7m]], [[5e7j]] – hDDX3X catalytic domain + nucleotide<br /> | ||
| - | **[[3dkp]] – hDDX52 domain I + ADP<BR /> | ||
| - | **[[2wax]], [[2way]] – hDDX6 C terminal + EDC3-FDF peptide<BR /> | ||
| - | **[[2j0s]], [[2j0q]], [[2hyi]] – hDDX48 + protein mago nashi homolog + RNA-binding protein 8A + protein CASC3 + RNA<BR /> | ||
| - | **[[2j0u]] - hDDX48 + protein CASC3<BR /> | ||
| - | **[[2db3]] – DmVASA residues 200-623 + RNA – ''Drosophila melanogaster''<br /> | ||
| - | **[[5nt7]] – DmVASA RecA-like domain + maternal effect protein<br /> | ||
| - | **[[3rrm]] – yDBP5 residues 91-482 (mutant) + GLE1 + NUP159 + ADP + IP6<BR /> | ||
| - | **[[3rrn]] - yDBP5 residues 91-482 (mutant) + GLE1 + IP6<BR /> | ||
| - | **[[5elx]] - yDBP5 residues 91-482 + RNA<br /> | ||
| - | **[[3pew]], [[3pey]] - yDBP5 residues 91-482 (mutant) + ADP + BeF3 + RNA<BR /> | ||
| - | **[[3peu]], [[3pev]] - yDBP5 C-terminal (mutant) + GLE1 + IP6<BR /> | ||
| - | **[[3i5x]], [[3i5y]] – yMSS116 + AMPPNP + RNA<BR /> | ||
| - | **[[3sqw]], [[3sqx]] - yMSS116 + AMPPNP + RNA<BR /> | ||
| - | **[[3i61]], [[4tyw]], [[4tyy]], [[4tz0]], [[4tz6]] - yMSS116 + nucleotide + BeF3 + RNA<BR /> | ||
| - | **[[3i62]] - yMSS116 + ADP + AlF4 + RNA<BR /> | ||
| - | **[[4tyn]] - yMSS116 + ADP + DNA<br /> | ||
| - | **[[2xau]], [[3kx2]], [[5jpt]] – yPrp43 + nucleotide<BR /> | ||
| - | **[[5i8q]] - yPrp43 + ADPNP + RNA<br /> | ||
| - | **[[5ltk]], [[5ltj]], [[5d0u]] - CtPrp43 + ADP – ''Chaetomium thermophilum''<br /> | ||
| - | **[[5lta]] - CtPrp43 + ADP + RNA<br /> | ||
| - | **[[3jrv]] – DDX3X + protein K7 – Vaccinia virus<BR /> | ||
| - | **[[3nbf]] - TtHera N terminal (mutant) + 8-oxo-ADP<br /> | ||
| - | **[[3moj]] - BsDBPA RNA-binding domain + RNA<br /> | ||
| - | **[[6aic]] – SaDB CSHA N-terminal + AMP <br /> | ||
| - | |||
| - | * DEAH box RNA helicase | ||
| - | |||
| - | **[[2eqs]] – hDHX8 S1 domain residues 260-355<br /> | ||
| - | **[[3i4u]] – hDHX8 residues 950-1183<br /> | ||
| - | **[[3llm]] – hDHX9<br /> | ||
| - | **[[5xdr]] – hDHX15 residues 110-795 (mutant)<br /> | ||
| - | **[[2n16]] – hDHX36 G-quadruplex domain - NMR<br /> | ||
| - | **[[2rqa]] – hDHX58 C terminal – NMR<br /> | ||
| - | **[[3vyx]], [[3vyy]] – hDHX9 RNA-binding domain + RNA<br /> | ||
| - | **[[2n21]] – hDHX36 G-quadruplex domain + DNA - NMR<br /> | ||
| - | **[[5e3h]] - hDHX58 residues 232-925 + RNA<br /> | ||
| - | **[[3eqt]]- hDHX58 residues 541-678 + RNA<br /> | ||
| - | **[[5vha]] – bDHX36 residues 150-1010 - bovine<br /> | ||
| - | **[[5vhc]], [[5vhd]] – bDHX36 residues 150-1010 + ADP<br /> | ||
| - | **[[5vhe]] – bDHX36 residues 56-1010 + DNA<br /> | ||
| - | **[[2rs6]], [[2rs7]] – mDHX9 RNA-binding domain – NMR<br /> | ||
| - | **[[6heg]], [[6eud]] – EcHRPB<br /> | ||
| - | |||
| - | *ATP-dependent RNA helicase | ||
| - | |||
| - | **[[2g9n]] - hEIF4A dead domain<br /> | ||
| - | **[[5zbz]] - hEIF4A dead domain + sanguinarine<br /> | ||
| - | **[[5zc9]] - hEIF4A dead domain + ATP + RNA<br /> | ||
| - | **[[6ieg]] - hDOB1 residues 70-1042 <br /> | ||
| - | **[[6ro1]] - hDOB1 residues 70-1042 + NVL<br /> | ||
| - | **[[6ieh]] - hDOB1 residues 70-1042 + NRDE2<br /> | ||
| - | **[[6c90]] - hDOB1 residues 70-600 + ZCCHC8<br /> | ||
| - | **[[6d6r]], [[6d6q]] - hDOB1 in exosome + RNA – Cryo EM<br /> | ||
| - | **[[2xzl]] – yNAM7 CH and helicase domains + RNA<BR /> | ||
| - | **[[2xgj]] – yDOB1 residues 81-1073 + RNA<BR /> | ||
| - | **[[4u4c]] - yDOB1 residues 81-1073 + poly(A) RNA polymerase protein 2<br /> | ||
| - | **[[4qu4]] - yDOB1 <br /> | ||
| - | **[[5ooq]] - yDOB1 residues 81-1073 + NOP53<br /> | ||
| - | **[[5suq]] – ySUB2 + TEX1 + THO2 <br /> | ||
| - | **[[5sup]] – ySUB2 + YRA1 + RNA <br /> | ||
| - | **[[2vso]], [[2vsx]] – yEIF4A + initiation factor 4F middle domain<BR /> | ||
| - | **[[2g9n]] - hEIF4A dead domain | ||
| - | |||
| - | *Viral ATP-dependent RNA helicase NS3 | ||
| - | |||
| - | **[[3o8b]], [[3o8d]], [[1cu1]], [[8ohm]], [[1hei]], [[4a92]], [[3rvb]] – HvHel NS3 residues 1186-1658 – Hepatitis C virus<BR /> | ||
| - | **[[1onb]], [[1jr6]] - HvHel NS3 arginine-rich domain residues 1353-1507 – NMR<BR /> | ||
| - | **[[2jlq]], [[2bhr]], [[2bmf]], [[2whx]], [[2wzq]] – DvHel NS3 residues 1646-2092 – Dengue virus<BR /> | ||
| - | **[[5xc7]] – DvHel NS3 residues 1646-2092 (mutant) <br /> | ||
| - | **[[2z83]] – Hel NS3 residues 167-624 – Japanese encephalitis virus<BR /> | ||
| - | **[[2v8o]], [[2wv9]] - Hel NS3 helicase domain – Murray valley encephalitis virus<BR /> | ||
| - | **[[2qeq]] - Hel NS3 residues 1691-2124 – Kunjin virus<BR /> | ||
| - | **[[5txg]], [[5jmt]], [[5vi7]], [[5jps]], [[6mh3]] - ZvHel NS3 helicase domain – Zika virus<br /> | ||
| - | **[[5k8u]], [[5k8l]], [[5k8i]], [[5jwh]], [[5jrz]] - ZvHel NS3 residues 164-621<br /> | ||
| - | **[[5x8y]] - ZvHel NS1 residues 168-352<br /> | ||
| - | **[[2v6i]] – KvHel helicase domain – Kokobera virus<BR /> | ||
| - | **[[2v6j]] - KvHel helicase domain (mutant)<br /> | ||
| - | **[[5wso]], [[5gvu]] - Hel NS3 residues 1939-2414 – bovine viral diarrhea virus<br /> | ||
| - | |||
| - | *''Viral ATP-dependent RNA helicase NS3 complexes'' | ||
| - | |||
| - | **[[3o8c]], [[3o8r]], [[3kqh]], [[3kqk]], [[3kql]], [[3kqn]], [[3kqu]], [[1a1v]] - HvHel NS3 + RNA<BR /> | ||
| - | **[[2f9v]] - HvHel NS3 protease domain + polyprotein<BR /> | ||
| - | **[[2a4g]], [[1rtl]], [[1w3c]], [[1dxp]] - HvHel NS3 protease domain + NS4A peptide<BR /> | ||
| - | **[[2a4q]], [[2a4r]], [[2f9u]], [[2fm2]], [[1dy8]], [[1dy9]] - HvHel NS3 protease domain + NS4A peptide + inhibitor<BR /> | ||
| - | **[[2jlr]] - DvHel NS3 residues 1646-2092 + AMPPNP<BR /> | ||
| - | **[[2jls]] - DvHel NS3 residues 1646-2092 + ADP<BR /> | ||
| - | **[[2jlu]], [[2jlw]], [[5xc6]] - DvHel NS3 residues 1646-2092 + RNA<BR /> | ||
| - | **[[2jlv]] - DvHel NS3 residues 1646-2092 + AMPPNP + RNA<BR /> | ||
| - | **[[2jlx]], [[2jly]], [[2jlz]] - DvHel NS3 residues 1646-2092 + ADP + RNA<BR /> | ||
| - | **[[2vbc]] - DvHel NS3 residues 1475-2092 + NS2A peptide<br /> | ||
| - | **[[5k8t]] - ZvHel NS3 residues 164-621 + GTP<br /> | ||
| - | **[[5gjc]] - ZvHel NS3 residues 1676-2119 + ATP<br /> | ||
| - | **[[5gjb]] - ZvHel NS3 residues 1676-2119 + RNA<br /> | ||
| - | **[[5y4z]] - ZvHel NS3 residues 1676-2119 + AMPPNP<br /> | ||
| - | |||
| - | *ATP-dependent RNA helicase SUV3 | ||
| - | |||
| - | **[[3rc3]] – hSUV3 residues 47-722<BR /> | ||
| - | **[[3og8]] – hDDX58 C terminal + RNA<BR /> | ||
| - | **[[3rc8]] - hSUV3 residues 47-722 + RNA<BR /> | ||
| - | **[[6f4a]] – ySUV3 + exoribonuclease II + RNA<br /> | ||
| - | |||
| - | *ATP-dependent RNA helicase A | ||
| - | |||
| - | **[[3llm]] – hHel A<BR /> | ||
| - | **[[3vyx]], [[3vyy]] - hHel A RNA-binding domain + RNA<br /> | ||
| - | **[[1whq]], [[2rs6]], [[2rs7]] – Hel A RNA-binding domain – mouse - NMR<BR /> | ||
| - | |||
| - | *ATP-dependent RNA helicase Repa | ||
| - | |||
| - | **[[1uaa]] – EcRepa + DNA<br /> | ||
| - | **[[1nlf]] - EcRepa + sulfate<br /> | ||
| - | **[[1g8y]], [[1olo]] - EcRepa | ||
| - | |||
| - | *ATP-dependent RNA helicase HEF | ||
| - | |||
| - | **[[1x2i]] – HEF DNA-binding domain – ''Pyrococcus furiosus'' | ||
| - | |||
| - | *ATP-dependent RNA helicase Srmb | ||
| - | |||
| - | **[[2yjt]] - EcSrmb residues 219-388 + RRAA<br /> | ||
| - | |||
| - | *ATP-dependent RNA helicase Dhh1 | ||
| - | |||
| - | **[[4bru]] – yDhh1 (mutant) + EDC3<br /> | ||
| - | **[[4brw]] – yDhh1 (mutant) + PAT1<br /> | ||
| - | |||
| - | *Werner syndrome ATP-dependent DNA helicase | ||
| - | |||
| - | **[[2e1e]], [[2e1f]], [[2dgz]] – hWRN HRDC domain<BR /> | ||
| - | **[[2fbt]] - hWRN exonuclease domain<BR /> | ||
| - | **[[2fbv]], [[2fbx]], [[2fby]], [[2fc0]] - hWRN exonuclease domain + metal ion | ||
| - | **[[3aaf]] – hWRN C terminal + DNA<br /> | ||
| - | **[[2e6l]], [[2e6m]]- mWRN exonuclease domain | ||
| - | |||
| - | *ATP-dependent DNA helicase Q | ||
| - | |||
| - | **[[2v1x]] - hQ1 residues 49-616<BR /> | ||
| - | **[[2wwy]], [[4u7d]] – hQ1 residues 49-616 + DNA<br /> | ||
| - | **[[5lst]] – hQ4 residues 427-1116<br /> | ||
| - | **[[2kmu]] – hQ4 N terminal - NMR<br /> | ||
| - | **[[5lb8]] – hQ5 <br /> | ||
| - | **[[5lb5]], [[5lb3]] – hQ5 + ADP<br /> | ||
| - | **[[5lba]] – hQ5 + urea derivative<br /> | ||
| - | |||
| - | *ATP-dependent DNA helicase RecG | ||
| - | |||
| - | **[[1gm5]] – TmRecG + DNA <BR /> | ||
| - | |||
| - | *ATP-dependent DNA helicase RecQ (Bloom Syndrome helicase) | ||
| - | |||
| - | **[[4cgz]], [[4o3m]] - hRecQ catalytic domain + DNA<br /> | ||
| - | **[[4cdg]] - hRecQ catalytic domain + nanobody<br /> | ||
| - | **[[1d8b]] - yRecQ HRDC domain - NMR<BR /> | ||
| - | **[[3iuo]] – RecQ residues 604-725 – ''Porphyromonas gingivalis''<BR /> | ||
| - | **[[2rhf]] – DrRecQ HRDC domain 3 – ''Deinococcus radiodurans''<BR /> | ||
| - | **[[2ma1]] - DrRecQ HRDC domain – NMR <br /> | ||
| - | **[[4q48]] - DrRecQ catalytic domain <br /> | ||
| - | **[[4q47]] - DrRecQ catalytic domain + ADP <br /> | ||
| - | **[[1wud]] - EcRecQ HRDC domain<BR /> | ||
| - | **[[1oyw]] - EcRecQ catalytic domain<BR /> | ||
| - | **[[1oyy]] - EcRecQ catalytic domain + ATP<BR /> | ||
| - | |||
| - | *ATP-dependent DNA helicase RuvA, RuvB | ||
| - | |||
| - | **[[2ztc]], [[2ztd]], [[2zte]], [[2h5x]] – MtRuvA – ''Mycobacterium tuberculosis''<BR /> | ||
| - | **[[1bvs]] - RuvA – ''Mycobacterium leprae''<BR /> | ||
| - | **[[1cuk]], [[1hjp]] – EcRuvA<br /> | ||
| - | **[[1d8l]] – EcRuvA N terminal<BR /> | ||
| - | **[[1c7y]], [[1bdx]] – EcRuvA + DNA<BR /> | ||
| - | **[[1hqc]] – TtRuvB<br /> | ||
| - | **[[1ixr]], [[1ixs]] – TtRuvA + RuvB (mutant) <BR /> | ||
| - | **[[1in4]] – TmRuvB<BR /> | ||
| - | **[[1in5]], [[1in6]], [[1in7]], [[1in8]], [[1j7k]] – TmRuvB (mutant)<br /> | ||
| - | **[[3pfi]] – RuvB + ADP – ''Campylobacter jejuni''<br /> | ||
| - | **[[6blb]] - PaRuvB + ADP - ''Pseudomonas aeruginosa''<br /> | ||
| - | |||
| - | *ATP-dependent DNA helicase UVSW | ||
| - | |||
| - | **[[2oca]], [[1rif]]- T4UVSW (mutant) – Enterobacteria phage T4<BR /> | ||
| - | **[[2jpn]] - T4UVSW | ||
| - | |||
| - | *ATP-dependent DNA helicase DDA | ||
| - | |||
| - | **[[3upu]] – T4DDA + DNA<br /> | ||
| - | |||
| - | *ATP-dependent DNA helicase PriA | ||
| - | |||
| - | **[[2d7e]] – EcPriA DNA-binding domain<BR /> | ||
| - | **[[2d7g]], [[2d7h]], [[2dwl]], [[2dwm]], [[2dwn]] – EcPriA N terminal + polynucleotide | ||
| - | |||
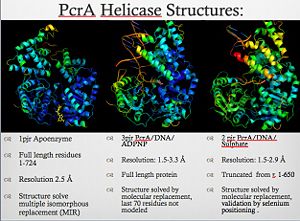
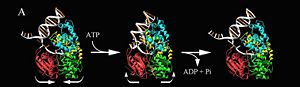
| - | *ATP-dependent DNA helicase PcrA | ||
| - | |||
| - | **[[1qhg]] – GsPcrA (mutant) + ADPNP <BR /> | ||
| - | **[[1qhh]] - GsPcrA + ADPNP<br /> | ||
| - | **[[2pjr]], [[3pjr]] - GsPcrA + DNA<br /> | ||
| - | **[[1pjr]] - GsPcrA<br /> | ||
| - | **[[5dma]] – GsPcrA C-terminal tudor domain<br /> | ||
| - | |||
| - | *DnaB helicase | ||
| - | |||
| - | **[[3gxv]] – HpDnaB – ''Helicobacter pylori''<BR /> | ||
| - | **[[4a1f]] – HpDnaB C terminal<br /> | ||
| - | **[[2r6d]] – BsDnaB – ''Bacillus stearothermophilus''<BR /> | ||
| - | **[[2r6e]], [[2r6d]] - GsDnaB <BR /> | ||
| - | **[[2r5u]] – MtDnaB N terminal<BR /> | ||
| - | **[[3bgw]] – BpDnaB-like – Bacillus phage SPP1<BR /> | ||
| - | **[[3bh0]] - BpDnaB-like G40P ATPase domain<BR /> | ||
| - | **[[2q6t]] – DnaB – ''Thermus aquaticus''<BR /> | ||
| - | **[[1mi8]], [[6frh]] – SyDnaB – ''Synechocystis''<BR /> | ||
| - | **[[6frg]], [[6fre]] – SyDnaB (mutant) <br /> | ||
| - | **[[6bbm]] – EcDnaB + replication protein P – Cryo EM<br /> | ||
| - | **[[1b79]] - EcDnaB N terminal<BR /> | ||
| - | **[[1jwe]] - EcDnaB N terminal - NMR<BR /> | ||
| - | **[[2r6c]], [[2r6a]] – BsDnaB + DnaG helicase-binding domain<br /> | ||
| - | **[[6bs8]] – MsDnaB <br /> | ||
| - | |||
| - | *DnaC helicase | ||
| - | |||
| - | **[[2vyf]]– GkDnaC – ''Geobacillus kaustophilus''<br /> | ||
| - | **[[2vye]] – GkDnaC + DNA <br /> | ||
| - | **[[3ec2]] – AaDnaC – ''Aquifex aeolicus''<br /> | ||
| - | **[[3ecc]] – AaDnaC + ADP<br /> | ||
| - | **[[6qel]] – EcDnaC + replicative DNA Hel <br /> | ||
| - | **[[6qem]] – EcDnaC + replicative DNA Hel + DNA<br /> | ||
| - | |||
| - | *Helicase BLM | ||
| - | |||
| - | **[[5lup]] – hBLM DHBN domain <br /> | ||
| - | **[[5lut]] – BLM DHBN domain - chicken<br /> | ||
| - | **[[5lus]] – BLM DHBN domain - pelican<br /> | ||
| - | |||
| - | *Helicase CA8 or Prp28 | ||
| - | |||
| - | **[[4w7s]] – yCa8<br /> | ||
| - | |||
| - | *Helicase NSP2 | ||
| - | |||
| - | **[[6jim]] – NSP2 + RNA – Chicungunya virus<br /> | ||
| - | **[[2hwk]] – NSP2 – Venzuelan equine encephalitis virus | ||
| - | |||
| - | *Helicase SEN1 | ||
| - | |||
| - | **[[5mzn]], [[6i59]] – ySEN1<br /> | ||
| - | |||
| - | *Helicase DING | ||
| - | |||
| - | **[[6fws]], [[6fwr]] – EcDING + DNA + ADP<br /> | ||
| - | |||
| - | *Helicase SNF2 | ||
| - | |||
| - | **[[1z5z]] – SsSNF2 C terminal – ''Sulfolobus solfataricus''<br /> | ||
| - | **[[1z63]] - SsSNF2 ATPase core + DNA<br /> | ||
| - | **[[1z6a]] - SsSNF2 ATPase core | ||
| - | |||
| - | *Helicase SKI2 | ||
| - | |||
| - | **[[4a4k]] – ySKI2 insertion domain<br /> | ||
| - | **[[4a4z]] – ySKI2 + AMPPNP<br /> | ||
| - | **[[2va8]] – SsSKI2<br /> | ||
| - | |||
| - | *Helicase SWR1 | ||
| - | |||
| - | **[[5i9e]] – ySWR1 + actin + actin-related protein<br /> | ||
| - | |||
| - | *Helicase XPD | ||
| - | |||
| - | **[[3crv]], [[3crw]], [[5h8c]] – XPD – ''Sulfolobus acidocaldarius''<br /> | ||
| - | **[[4a15]] - TaXPD + DNA <br /> | ||
| - | **[[2vsf]] - TaXPD<br /> | ||
| - | **[[2vl7]] - XPD - ''Sulfolobus tokodaii''<br /> | ||
| - | **[[5ivw]] - hXPD + XPB + TFIIH ubunits 2,3,4,5 + SCP-X + SCP-Ybr /> | ||
| - | |||
| - | *Pre-mRNA-splicing Helicase | ||
| - | |||
| - | **[[3hib]], [[3im1]], [[3im2]] – yBrr2 2nd SEC63 domain<br /> | ||
| - | **[[4bgd]], [[5m5p]], [[5m52]], [[5dca]] – yBrr2 + pre-mRNA-splicing factor 8<br /> | ||
| - | |||
| - | *crispr-associated helicase Cas3 | ||
| - | |||
| - | **[[4q2c]] – TteCas3 – ''Thermobaculum terrenum''<br /> | ||
| - | **[[4q2d]] – TteCas3 + DTP<br /> | ||
| - | **[[4qqw]], [[4qqx]] – TfCas3 – ''Thermobifida fusca''<br /> | ||
| - | **[[4qqy]] – TfCas3 + ADP + HD nuclease<br /> | ||
| - | **[[4qqz]] – TfCas3 + AMPPNP <br /> | ||
| - | **[[6c66]] – TfCas3 + Cse1 + Cse3 + Cse4 + Cas5e – Cryo EM <br /> | ||
| - | **[[5gqh]] – PaCas3 + anti-crispr protein 3 <br /> | ||
| - | **[[5b7i]] – PaCas3 + ACRF3<br /> | ||
| - | |||
| - | }} | ||
==References== | ==References== | ||
Crystal structure of a DExx box DNA helicase., Subramanya HS, Bird LE, Brannigan JA, Wigley DB, Nature. 1996 Nov 28;384(6607):379-83. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/8934527 8934527]<br /> | Crystal structure of a DExx box DNA helicase., Subramanya HS, Bird LE, Brannigan JA, Wigley DB, Nature. 1996 Nov 28;384(6607):379-83. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/8934527 8934527]<br /> | ||
Revision as of 10:15, 23 July 2019
| |||||||||||
References
Crystal structure of a DExx box DNA helicase., Subramanya HS, Bird LE, Brannigan JA, Wigley DB, Nature. 1996 Nov 28;384(6607):379-83. PMID:8934527
^ Johnson DS, Bai L, Smith BY, Patel SS, Wang MD (2007). "Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped t7 helicase". Cell 129 (7): 1299–309. doi:10.1016/j.cell.2007.04.038. PMID 17604719.
^ a b "Researchers solve mystery of how DNA strands separate" (2007-07-03). Retrieved on 2007-07-05.
^ Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco Jr I, Pylr AM, Bustamante C, "RNA Translocation and Unwinding Mechanism of HCV NS3 Helicase and its Coordination by ATP", Nature. 2006 Jan 5; 439: 105-108.
Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for displacement of RecA protein from DNA or inhibition of RecA-mediated DNA strand exchange. Journal of Bacteriology (2007) 189 (12):4502-4509.
Bird L, Subramanya HS, Wigley DB, "Helicases: a unifying structural theme?", Current Opinion in Structural Biology. 1998 Feb; 8 (1): 14-18.
Betterton MD, Julicher F, "Opening of nucleic-acid double strands by helicases: active versus passive opening.", Physical Review E. 2005 Jan; 71 (1): 011904.
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006 Apr 21;125(2):287-300. PMID:16630817 doi:10.1016/j.cell.2006.01.054
- Sengoku T, Nureki O, Dohmae N, Nakamura A, Yokoyama S. Crystallization and preliminary X-ray analysis of the helicase domains of Vasa complexed with RNA and an ATP analogue. Acta Crystallogr D Biol Crystallogr. 2004 Feb;60(Pt 2):320-2. Epub 2004, Jan 23. PMID:14747711 doi:10.1107/S0907444903025897
Content Donators
Created with the participation of Luis E Ramirez-Tapia, Wayne Decatur.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Wayne Decatur, Joel L. Sussman