Pyrroline-5-carboxylate dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
== Structural highlights == | == Structural highlights == | ||
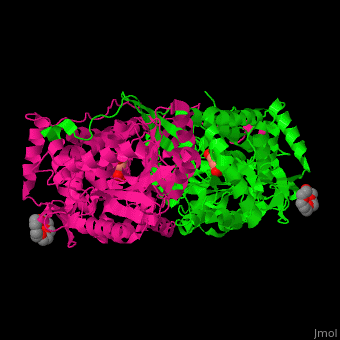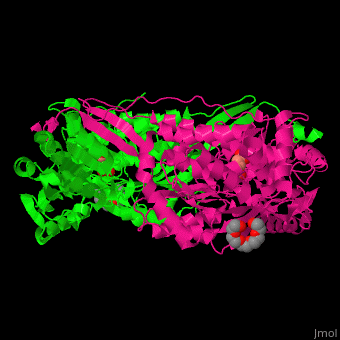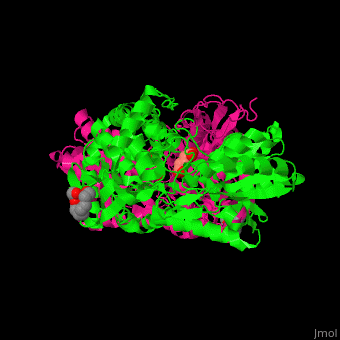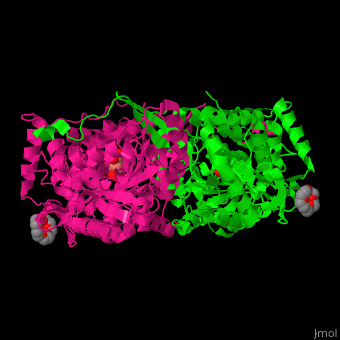
| - | PCD ligand <scene name='51/510199/Cv1/ | + | PCD ligand <scene name='51/510199/Cv1/4'>glutarate binds in the cleft between the catalytic and NAD-binding domains</scene>. Water molecules are shown as red spheres. A <scene name='51/510199/Cv1/5'>cystein residue is the nucleophile attacker of the aldehyde</scene><ref>PMID:23928095</ref>. |
</StructureSection> | </StructureSection> | ||
==3D structures of pyrroline-5-carboxylate dehydrogenase== | ==3D structures of pyrroline-5-carboxylate dehydrogenase== | ||
Revision as of 12:57, 21 August 2019
| |||||||||||
3D structures of pyrroline-5-carboxylate dehydrogenase
Updated on 21-August-2019
References
- ↑ Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004 Dec;16(12):3413-25. Epub 2004 Nov 17. PMID:15548746 doi:http://dx.doi.org/10.1105/tpc.104.023622
- ↑ Geraghty MT, Vaughn D, Nicholson AJ, Lin WW, Jimenez-Sanchez G, Obie C, Flynn MP, Valle D, Hu CA. Mutations in the Delta1-pyrroline 5-carboxylate dehydrogenase gene cause type II hyperprolinemia. Hum Mol Genet. 1998 Sep;7(9):1411-5. PMID:9700195
- ↑ Pemberton TA, Tanner JJ. Structural basis of substrate selectivity of Delta-pyrroline-5-carboxylate dehydrogenase (ALDH4A1): Semialdehyde chain length. Arch Biochem Biophys. 2013 Aug 6. pii: S0003-9861(13)00231-2. doi:, 10.1016/j.abb.2013.07.024. PMID:23928095 doi:10.1016/j.abb.2013.07.024