This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1111
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Generalities == | == Generalities == | ||
| - | The structure <scene name='82/829364/1psr/1'>1PSR</scene> is found in the human psoriasin, also called [https://en.wikipedia.org/wiki/S100A7 S100A7]. This protein belongs to the family of [http://proteopedia.org/wiki/index.php/Psoriasin S100] proteins. They are called like this because of their solubility in a saturated solution with ammonium sulfate at neutral pH <ref> Sedaghat F, Notopoulos A (October 2008). "S100 protein family and its application in clinical practice". ''Hippokratia''. '''4''': 198-204. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2580040/pdf/hippokratia-12-198.pdf </ref>. It is a family of 21 proteins of low molecular weights with a high frequence of homologous sequence (often between 40% and 50%). Those proteins are found in cells as homo and heterodimers. Each monomer have a similar structure, beginning with the N-terminal EF Hand and the four-turn <scene name='82/829364/Helix_1/2'>Helix I</scene> which leads to a <scene name='82/829364/Loop/1'>loop</scene>. Then, there is the three-turn <scene name='82/829364/Helix_ii/1'>Helix II</scene>, the two-turn <scene name='82/829364/Helix_iii/1'>Helix III</scene> and finally the five-turn <scene name='82/829364/Helix_iv/1'>Helix IV</scene>. One of their main properties is their ability to bind the calcium. They share some common structures such as two helix-loop-helix structures which are calcium-binding domains more specifically the loop from <scene name='82/829364/Calcium_binding_site/1'>Asp62 to Asp70</scene>. All the S100 proteins have different functions in many various cell types. They have significant roles in calcium-associated signal transduction. They play the roles of calcium sensors proteins that regulate the function or distribution of specific target proteins <ref>Eckert R, Broome AM, Ruse M, Robinson N, Ryan D, Lee K (July 2004). "S100 Proteins in the Epidermis". ''Journal of Investigative Dermatology''. '''123'''(1): 23-33.https://doi.org/10.1111/j.0022-202X.2004.22719.x</ref>. | + | The structure <scene name='82/829364/1psr/1'>1PSR</scene> is found in the human psoriasin, also called [https://en.wikipedia.org/wiki/S100A7 S100A7]. This protein belongs to the family of [http://proteopedia.org/wiki/index.php/Psoriasin S100] proteins whitch are involed in the skin celle division and differenciation. They are called like this because of their solubility in a saturated solution with ammonium sulfate at neutral pH <ref> Sedaghat F, Notopoulos A (October 2008). "S100 protein family and its application in clinical practice". ''Hippokratia''. '''4''': 198-204. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2580040/pdf/hippokratia-12-198.pdf </ref>. It is a family of 21 proteins of low molecular weights with a high frequence of homologous sequence (often between 40% and 50%). Those proteins are found in cells as homo and heterodimers. Each monomer have a similar structure, beginning with the N-terminal EF Hand and the four-turn <scene name='82/829364/Helix_1/2'>Helix I</scene> which leads to a <scene name='82/829364/Loop/1'>loop</scene>. Then, there is the three-turn <scene name='82/829364/Helix_ii/1'>Helix II</scene>, the two-turn <scene name='82/829364/Helix_iii/1'>Helix III</scene> and finally the five-turn <scene name='82/829364/Helix_iv/1'>Helix IV</scene>. One of their main properties is their ability to bind the calcium. They share some common structures such as two helix-loop-helix structures which are calcium-binding domains more specifically the loop from <scene name='82/829364/Calcium_binding_site/1'>Asp62 to Asp70</scene>. All the S100 proteins have different functions in many various cell types. They have significant roles in calcium-associated signal transduction. They play the roles of calcium sensors proteins that regulate the function or distribution of specific target proteins <ref>Eckert R, Broome AM, Ruse M, Robinson N, Ryan D, Lee K (July 2004). "S100 Proteins in the Epidermis". ''Journal of Investigative Dermatology''. '''123'''(1): 23-33.https://doi.org/10.1111/j.0022-202X.2004.22719.x</ref>. |
== Human Psoriasin == | == Human Psoriasin == | ||
Revision as of 21:40, 17 January 2020
| This Sandbox is Reserved from 25/11/2019, through 30/9/2020 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1091 through Sandbox Reserved 1115. |
To get started:
More help: Help:Editing |
1 PSR
| |||||||||||
References
- ↑ Sedaghat F, Notopoulos A (October 2008). "S100 protein family and its application in clinical practice". Hippokratia. 4: 198-204. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2580040/pdf/hippokratia-12-198.pdf
- ↑ Eckert R, Broome AM, Ruse M, Robinson N, Ryan D, Lee K (July 2004). "S100 Proteins in the Epidermis". Journal of Investigative Dermatology. 123(1): 23-33.https://doi.org/10.1111/j.0022-202X.2004.22719.x
- ↑ Hagens G, Masouyé I, Augsburger E, Hotz R, Saurat JH, Siegenthaler G (April 1999) "Calcium-binding protein S100A7 and epidermal-type fatty acid-binding protein are associated in the cytosol of human keratinocytes" Biochem J. 339(2): 419-427: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1220173/
- ↑ Chmurzyńska A (March 2006). "The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism".Journal of Applied Genetics.47(1): 39-48.https://link.springer.com/article/10.1007/BF03194597
- ↑ Brodersen DE, Etzerodt M, Madsen P, Celis JE, Thøgersen HC, Nyborg J, Kjeldgaard M (April 1998). "EF-hands at atomic resolution: the structure of human psoriasin(S100A7) solved by MAD phasing". Structure.6: 477-489.https://www.cell.com/structure/pdf/S0969-2126(98)00049-5.pdf
- ↑ Murray J, Boulanger M (April 2010). "S100A7 (S100 calcium binding protein A7)". Atlas of Genetics and Cytogeneticsin Oncology and Haematology. 15(1): 59-64. : http://AtlasGeneticsOncology.org/Genes/S100A7ID42194ch1q21.html
- ↑ S Alowami, G Qing, E Emberley, L Snell & PH Watson (2003). "Psoriasin , (S100A7) expression is altered during skin tumorigenesis" BMC Dermatology.3(1):
- ↑ AK Ekman, J Vegfors, C Bivik Eding, C Enerbäck (December 2016). "Overexpression of Psoriasin (S100A7) Contributes to Dysregulated Differentiation in Psoriasis", Acta Derm Venereol. 97: https://www.medicaljournals.se/acta/content/html/10.2340/00015555-2596
- ↑ KC Lee , RL Eckert (April 2007). "S100A7 (Psoriasin) – Mechanism of Antibacterial Action in Wounds", Journal of Investigative Dermatology 127(4): 945-957 https://www.sciencedirect.com/science/article/pii/S0022202X15333145
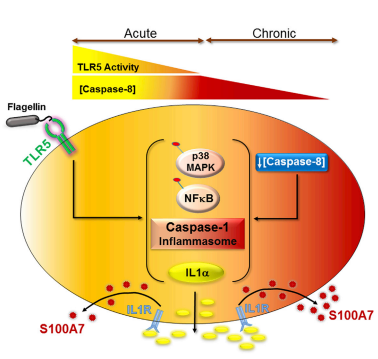
- ↑ T Bhatt, A Bhosale, B Bajantri, MS Mathapathi, A Rizvi, G Scita, A Majumdar and C Jamora (November 2019) "Sustained Secretion of the Antimicrobial Peptide S100A7 Is Dependent on the Downregulation of Caspase-8", Cell Reports. 29(9): 2546-2555 https://doi.org/10.1016/j.celrep.2019.10.090