UDP-galactopyranose mutase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
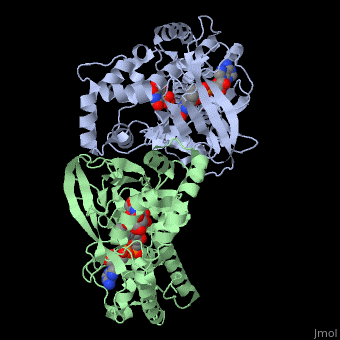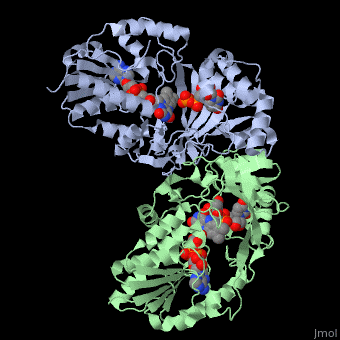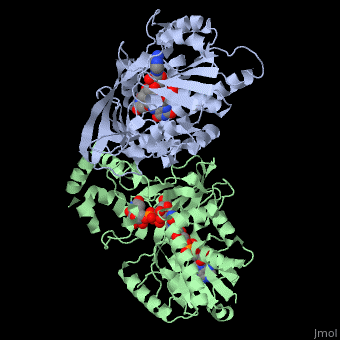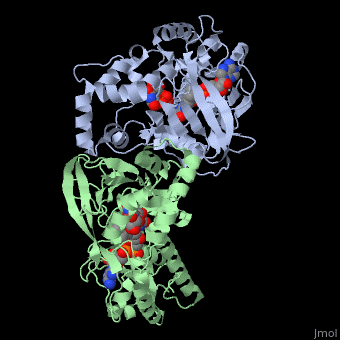
<StructureSection load='3int' size='340' side='right' caption='UDP-galactopyranose mutase dimer complex with FAD, uridine diphosphate, UDP-galactopyranose [[3int]]' scene='' > | <StructureSection load='3int' size='340' side='right' caption='UDP-galactopyranose mutase dimer complex with FAD, uridine diphosphate, UDP-galactopyranose [[3int]]' scene='' > | ||
| + | __TOC__ | ||
| + | ==Function== | ||
| + | |||
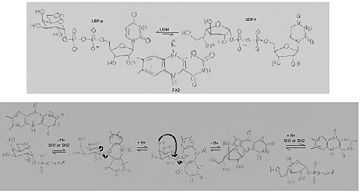
'''UDP-galactopyranose mutase''', UDP-D-Galactopyranose furanomutase<ref name="BEO1">http://www.brenda-enzymes.org/php/result_flat.php4?ecno=5.4.99.9</ref> or flavoenzyme uridine 5′-diphosphate galactopyranose mutase (UGM)<ref Name="GWF2">Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray Crystallography Reveals a Reduced Substrate Complex of UDP-Galactopyranose Mutase Poised for Covalent Catalysis by Flavin. Biochemistry. 2009 Oct 6; 48(39): 9171-73. [http://www.ncbi.nlm.nih.gov/pubmed/19719175 PMID:19719175]</ref>. | '''UDP-galactopyranose mutase''', UDP-D-Galactopyranose furanomutase<ref name="BEO1">http://www.brenda-enzymes.org/php/result_flat.php4?ecno=5.4.99.9</ref> or flavoenzyme uridine 5′-diphosphate galactopyranose mutase (UGM)<ref Name="GWF2">Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray Crystallography Reveals a Reduced Substrate Complex of UDP-Galactopyranose Mutase Poised for Covalent Catalysis by Flavin. Biochemistry. 2009 Oct 6; 48(39): 9171-73. [http://www.ncbi.nlm.nih.gov/pubmed/19719175 PMID:19719175]</ref>. | ||
It has a registry number of [[EC Number|EC]] 5.4.99.9<ref name="BEO1"/><ref name="MFD3">http://www.mondofacto.com/facts/dictionary?UDP-galactopyranose+mutase</ref>; and is an isomerase, (L) polypeptide that consists of a dimer made up of 2 monomers, a homodimer<ref name="PPE4">[http://www.pdb.org/pdb/explore/explore.do?structureId=3INT 3int RCSB PDB]</ref><ref name="BLB5">Beis K, Srikannathasan V, Liu H, Fullerton SWB, Bamford VA, Sanders DAR, Whitfield C, McNeil MR, Naismith JH. Crystal Structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the Oxidized State and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the (Active) Reduced State. J. Mol. Biol. 2005 May 13; 384(4): 971-982[http://www.ncbi.nlm.nih.gov/pubmed/15843027 PMID:15843027]</ref><ref name="GWK6">Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand Binding and Substrate Discrimination by UDP-Galactopyranose Mutase. J. Mol. Biol. 2009 Aug 14; 391(2): 327-340. [http://www.ncbi.nlm.nih.gov/pubmed/19500588 PMID:19500588]</ref>. | It has a registry number of [[EC Number|EC]] 5.4.99.9<ref name="BEO1"/><ref name="MFD3">http://www.mondofacto.com/facts/dictionary?UDP-galactopyranose+mutase</ref>; and is an isomerase, (L) polypeptide that consists of a dimer made up of 2 monomers, a homodimer<ref name="PPE4">[http://www.pdb.org/pdb/explore/explore.do?structureId=3INT 3int RCSB PDB]</ref><ref name="BLB5">Beis K, Srikannathasan V, Liu H, Fullerton SWB, Bamford VA, Sanders DAR, Whitfield C, McNeil MR, Naismith JH. Crystal Structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the Oxidized State and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the (Active) Reduced State. J. Mol. Biol. 2005 May 13; 384(4): 971-982[http://www.ncbi.nlm.nih.gov/pubmed/15843027 PMID:15843027]</ref><ref name="GWK6">Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand Binding and Substrate Discrimination by UDP-Galactopyranose Mutase. J. Mol. Biol. 2009 Aug 14; 391(2): 327-340. [http://www.ncbi.nlm.nih.gov/pubmed/19500588 PMID:19500588]</ref>. | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 http://www.brenda-enzymes.org/php/result_flat.php4?ecno=5.4.99.9
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray Crystallography Reveals a Reduced Substrate Complex of UDP-Galactopyranose Mutase Poised for Covalent Catalysis by Flavin. Biochemistry. 2009 Oct 6; 48(39): 9171-73. PMID:19719175
- ↑ 3.0 3.1 http://www.mondofacto.com/facts/dictionary?UDP-galactopyranose+mutase
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 3int RCSB PDB
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Beis K, Srikannathasan V, Liu H, Fullerton SWB, Bamford VA, Sanders DAR, Whitfield C, McNeil MR, Naismith JH. Crystal Structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the Oxidized State and Klebsiella pneumoniae UPD-Galactopyranose Mutase in the (Active) Reduced State. J. Mol. Biol. 2005 May 13; 384(4): 971-982PMID:15843027
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 6.12 6.13 6.14 6.15 Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand Binding and Substrate Discrimination by UDP-Galactopyranose Mutase. J. Mol. Biol. 2009 Aug 14; 391(2): 327-340. PMID:19500588
- ↑ 7.0 7.1 7.2 7.3 7.4 Zhang Q, Lui HW. Studies of UDP-Galactopyranose Mutase from Escherichia coli: An Unusual Role of Reduced FAD in its Cataysis. J. Am. Chem. Soc. 2000 Sep 27;122(38): 9065-70. DOI: 10.1021/ja001333z
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 Yao X, Bleile DW, Yuan Y, Chao J, Sarathy KP, Sanders DAR, Pinto BM, O’Neill MA. Substrate Directs Enzyme Dynamics by Bridging Distal Sites: UPD-Galactopyranose Mutase. Proteins: Structure, Function, Bioinformatics. 2008 June 30; 74(4): 972-79. PMID:18767162
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Joel L. Sussman, Christine Brown, Andrea Gorrell, Alexander Berchansky